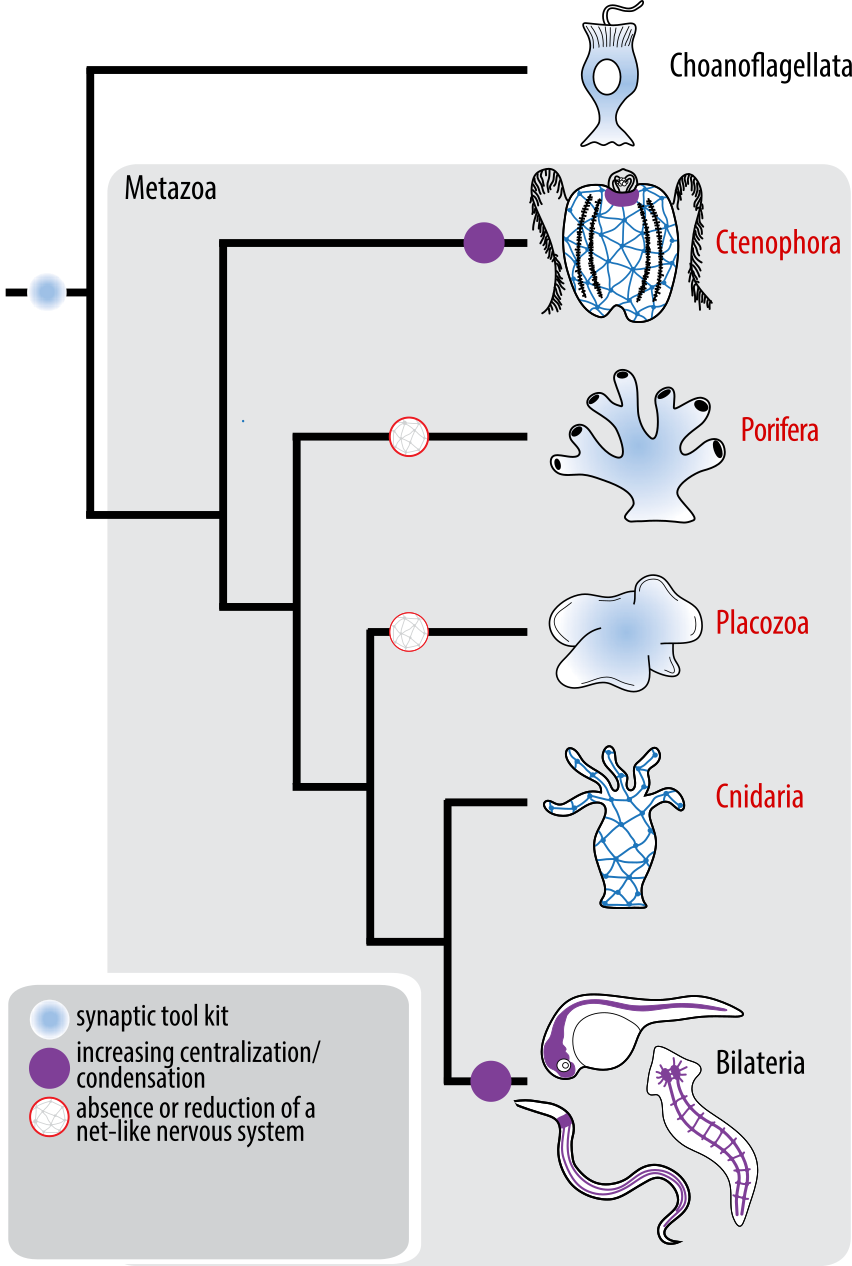
On chemical and synaptic brains and the evolution of nervous systems
School of Ideas in Neuroscience
July 15, 2024
Gáspár Jékely
Centre for Organismal Studies, Heidelberg University
What is a nervous system?
What is a nervous system?
- consists of cells
- neurons, glia etc.
- neurons are excitable
- neurons can influence each others activity
- and the activity of effectors
- some neurons may be sensory, tuned to environmental cues
- neurons have projections
- connections by synapses
- connections are specific (wiring specificity)
- organised into a complex network (connectome)
- is there a different way to wire complex networks?
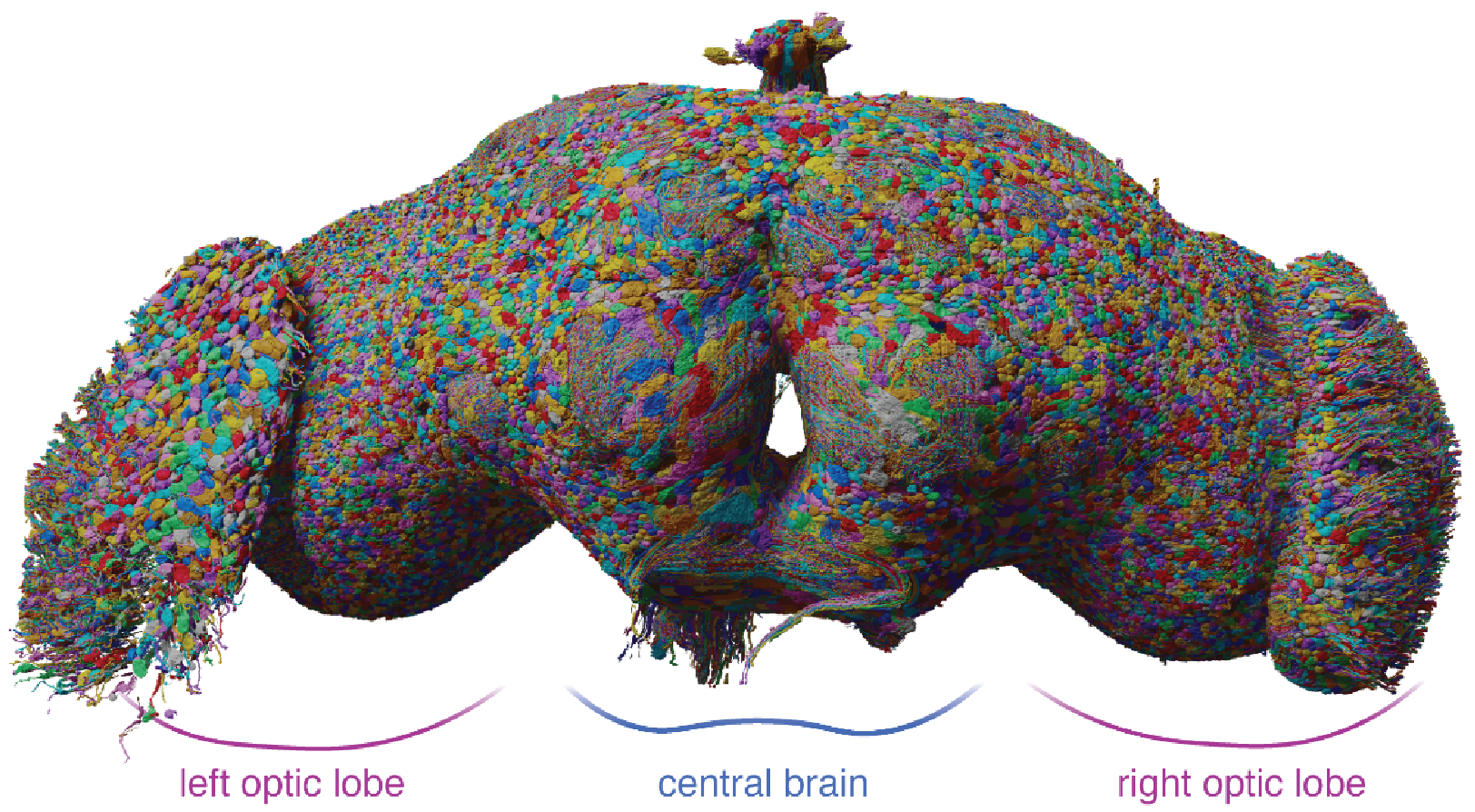
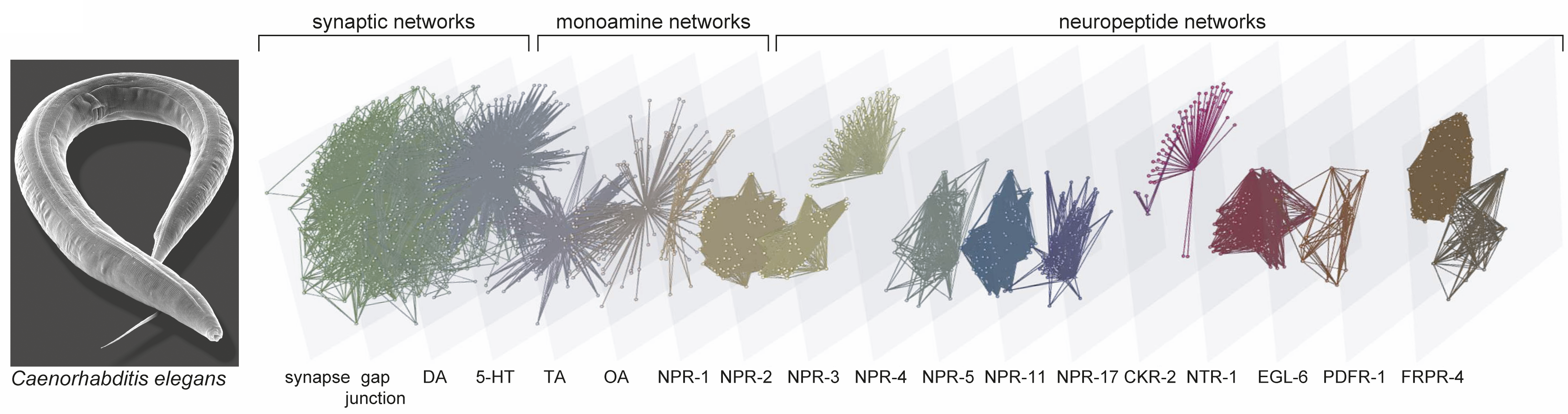
A synaptic connectivity matrix
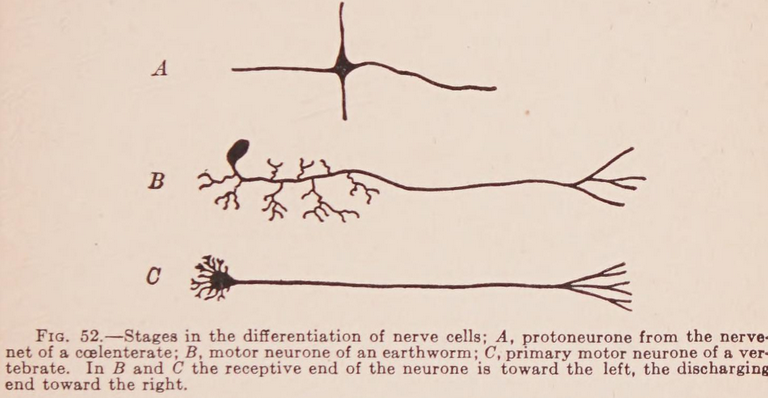
Origin of the nervous system as a reflex arc?

 George Howard Parker (1864 - 1955)
George Howard Parker (1864 - 1955)
Origin of the nervous system as a reflex arc?
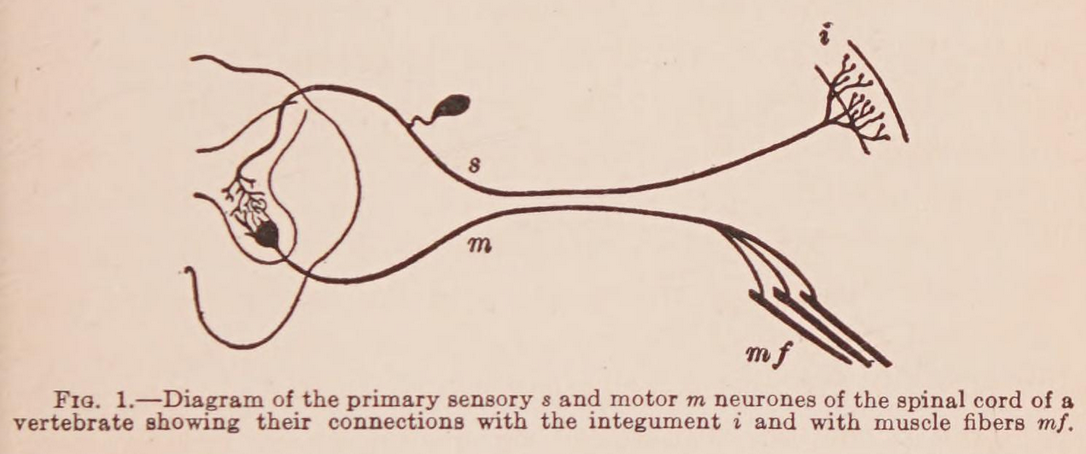
- independent effectors evolved first
- then receptor-effector systems
- then organisation into nerve nets
- diffuseness of transmission (no centralisation)
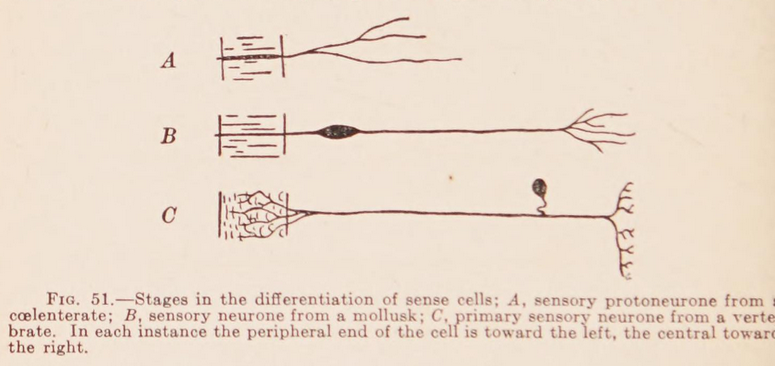
Neuronal communication by chemical signals?
- argued against the “connectionist view”
- proposed instead that neurons could communicate by specific chemical signals
- like a radio broadcast
- chemicals diffuse through the nervous system
- detected by specific chemical receptors in the target cells
- communications could occur in the absence of synaptic transmission
- discovery of pituitary hormones (1950ies)
 Paul Alfred Weiss (1898 – 1989)
Paul Alfred Weiss (1898 – 1989)
Chemical signalling between cells
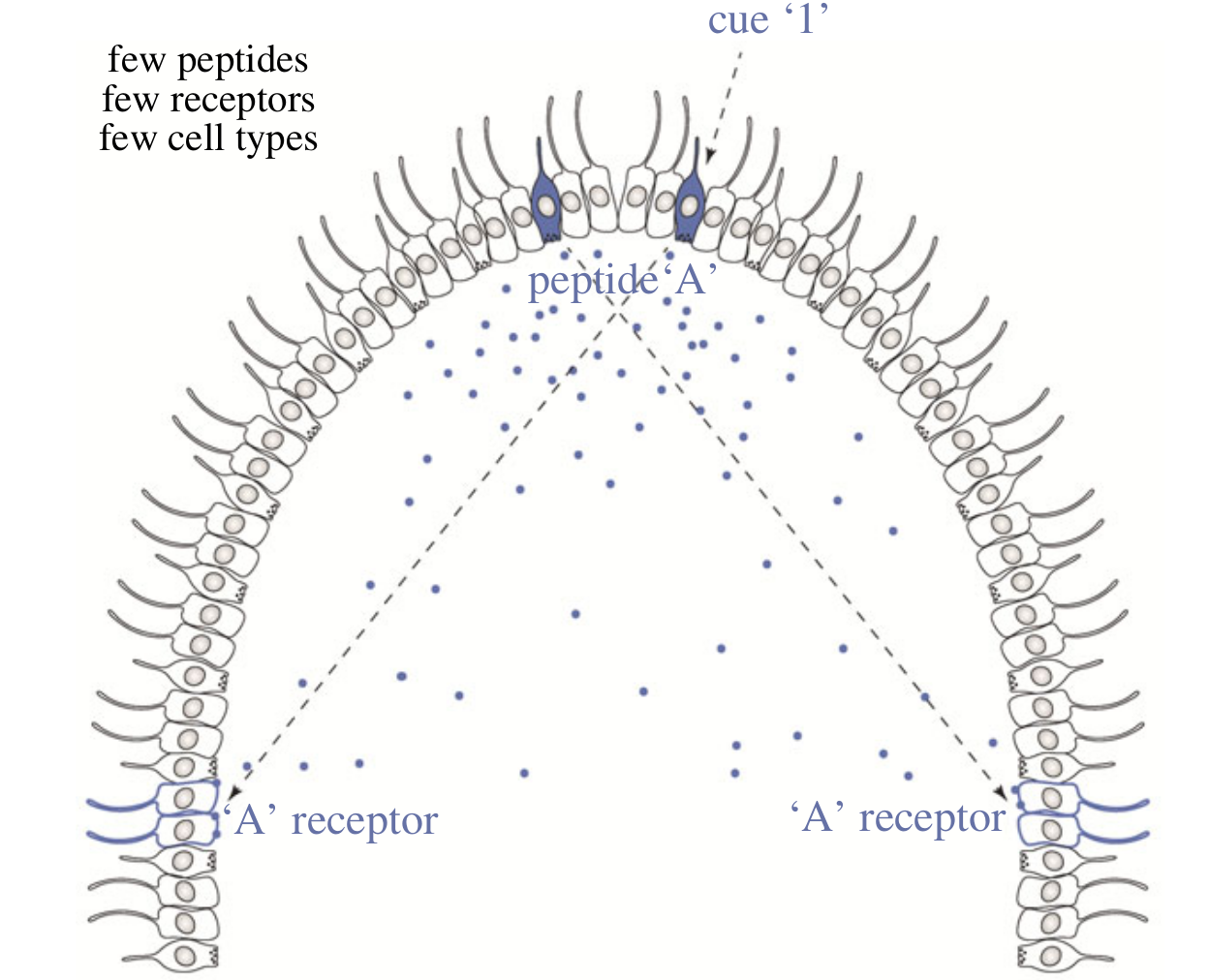
- some cells release a chemical (e.g. short peptide)
- other cells express a specific receptor for the chemical
- receptor activation -> change in cell state
- a specific cell-to-cell signalling is possible
- no synaptic connection
Chemical signalling between cells
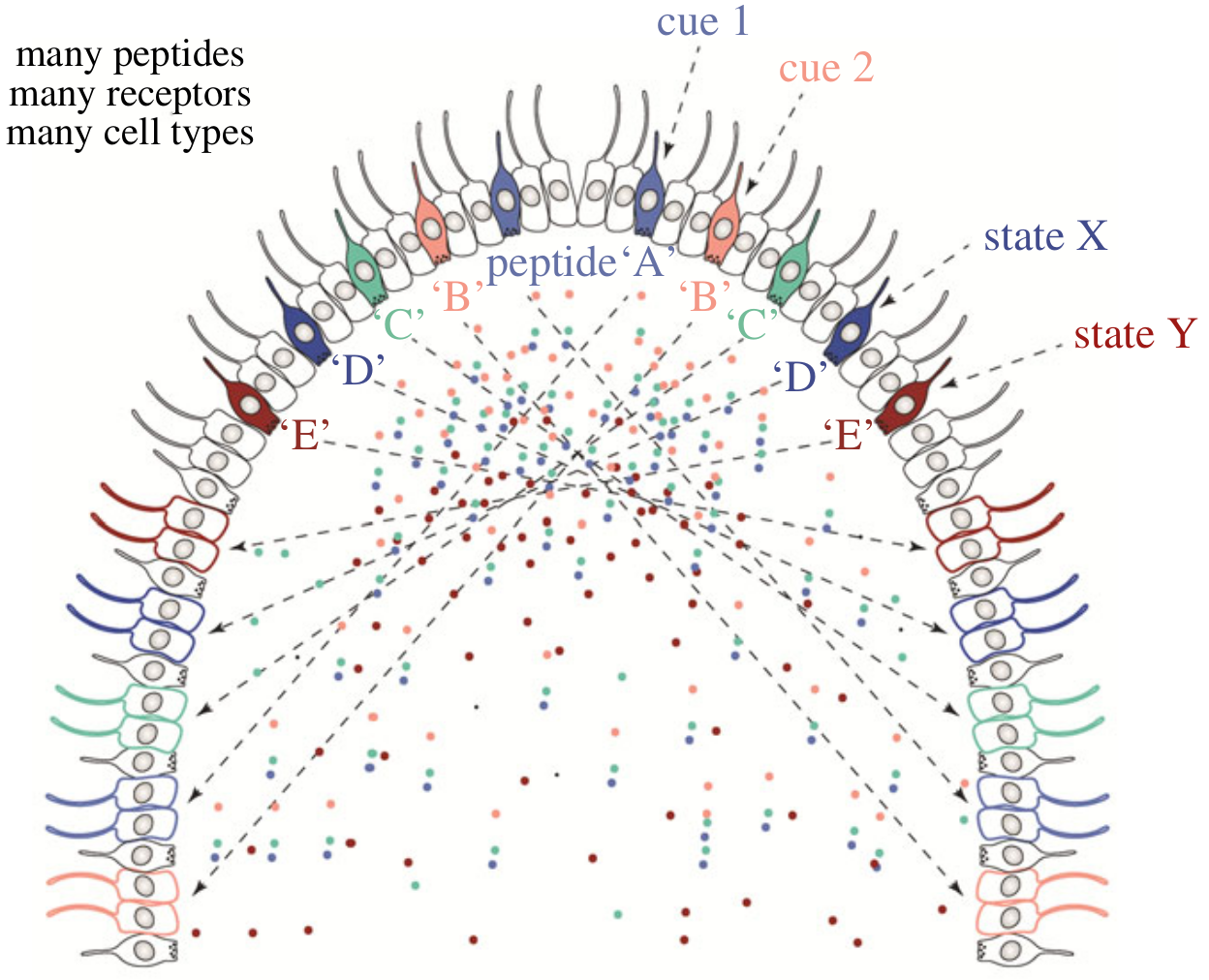
- several cell types, each expressing a different chemical signal
- several specific receptors expressed in target cells
- chemical ‘wiring diagram’
- no synaptic connection
Chemical connectomes
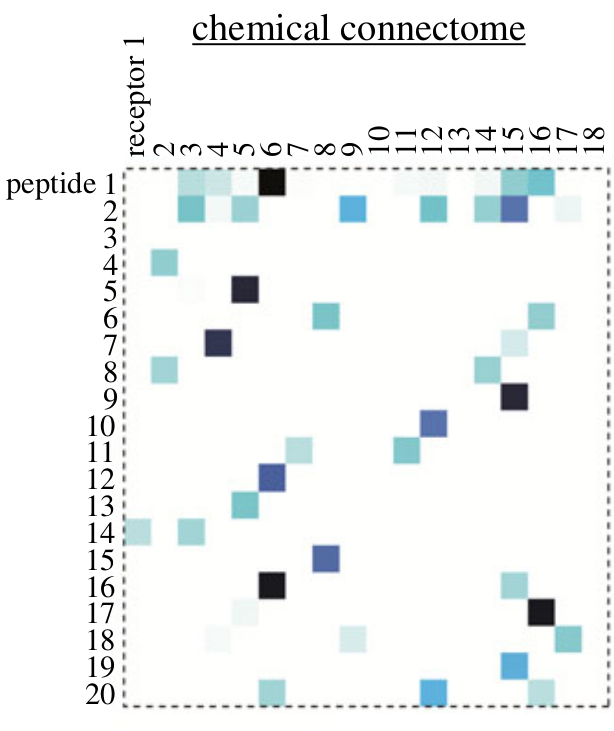
- sending and receiving cells
- a chemical connectivity matrix
- can be arbitrarily complex
Chemical connectomes
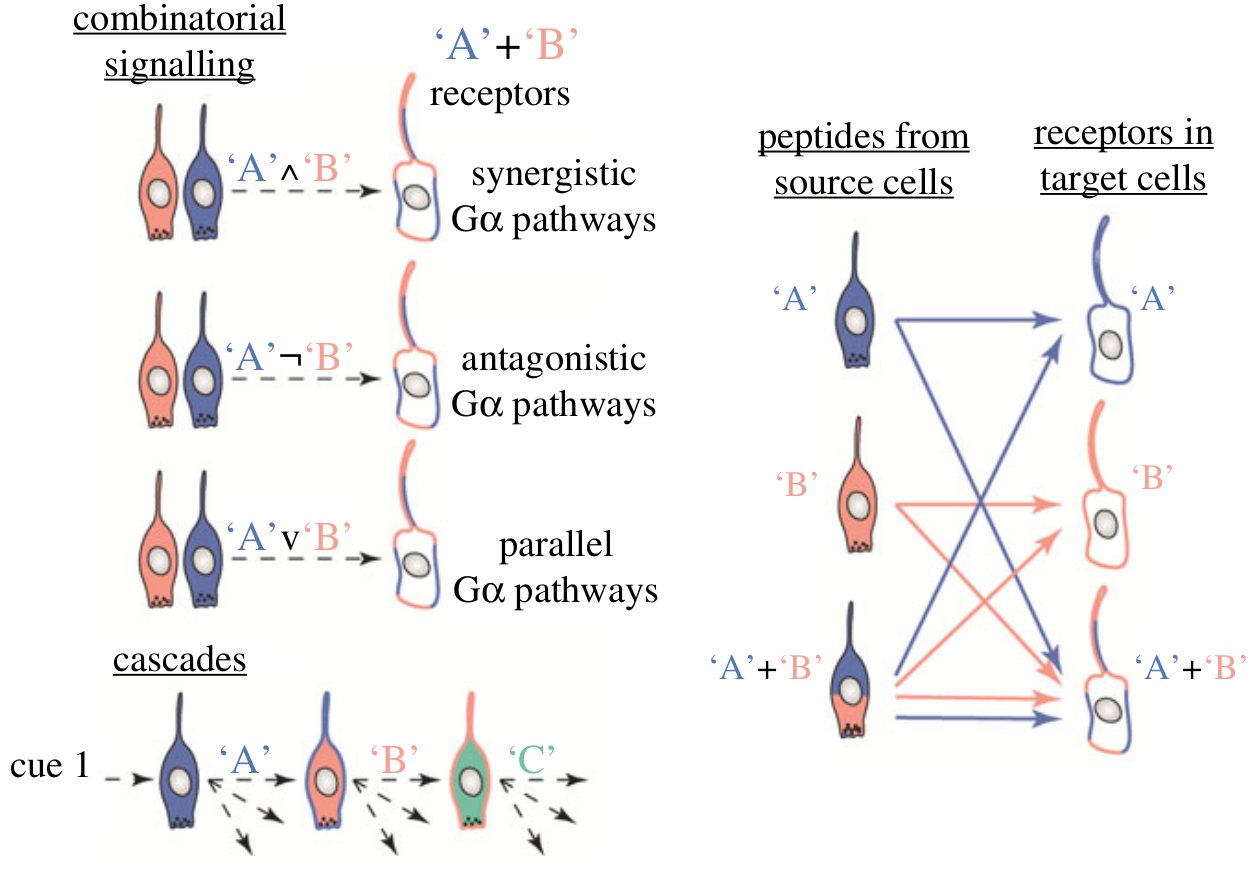
- co-expression of chemical signals or receptors
- combinatorics, cascades
- synergism, antagonism
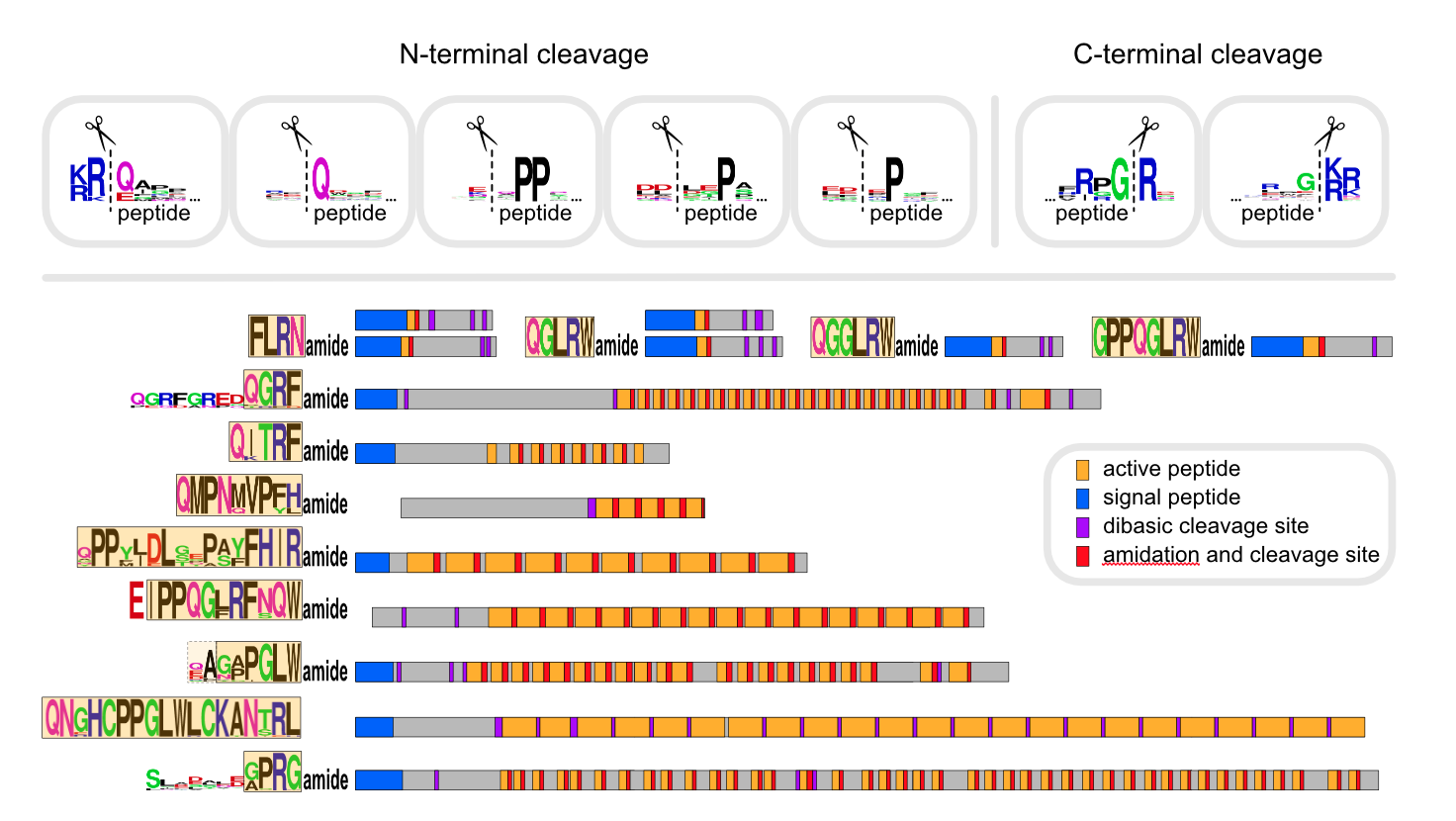
Neuropeptides are produced from precursor peptides

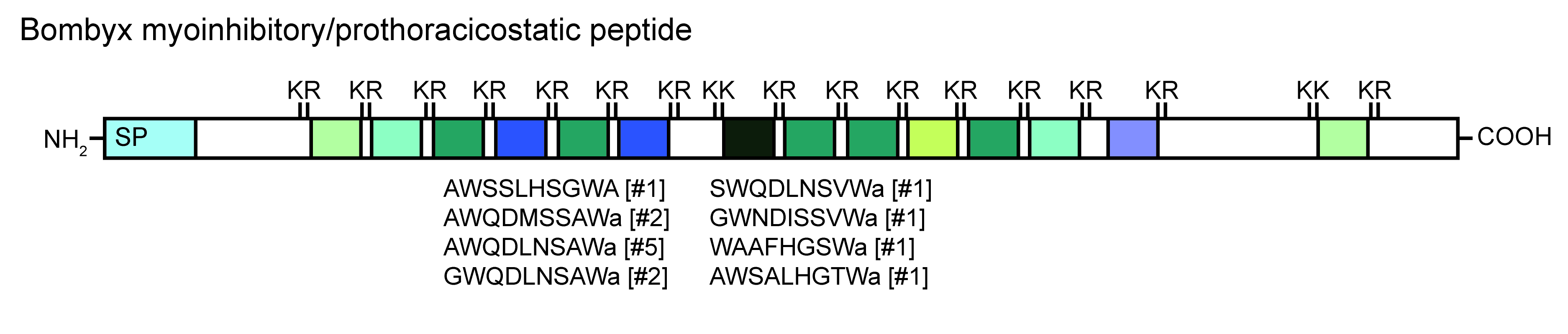
Neuropeptides are produced from precursor peptides
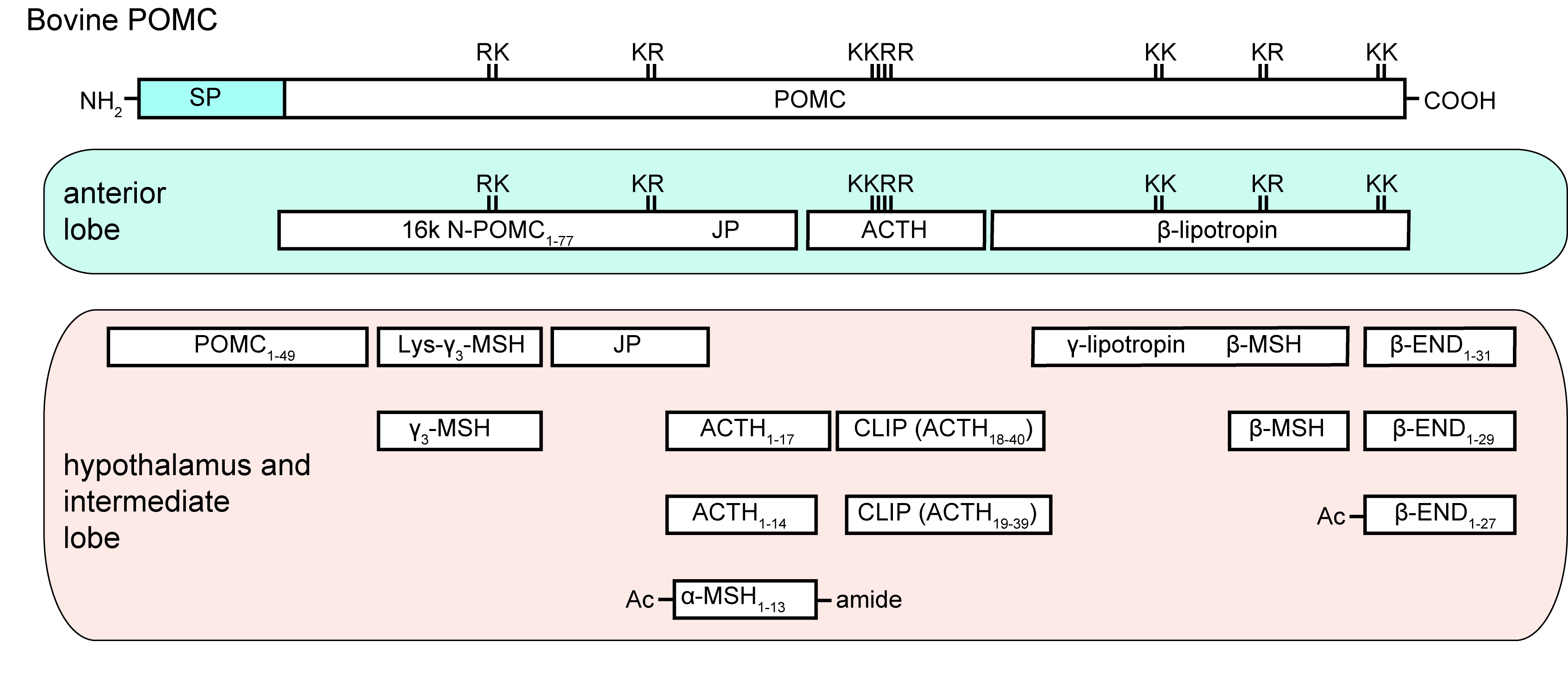
Neuropeptides are produced from precursor peptides
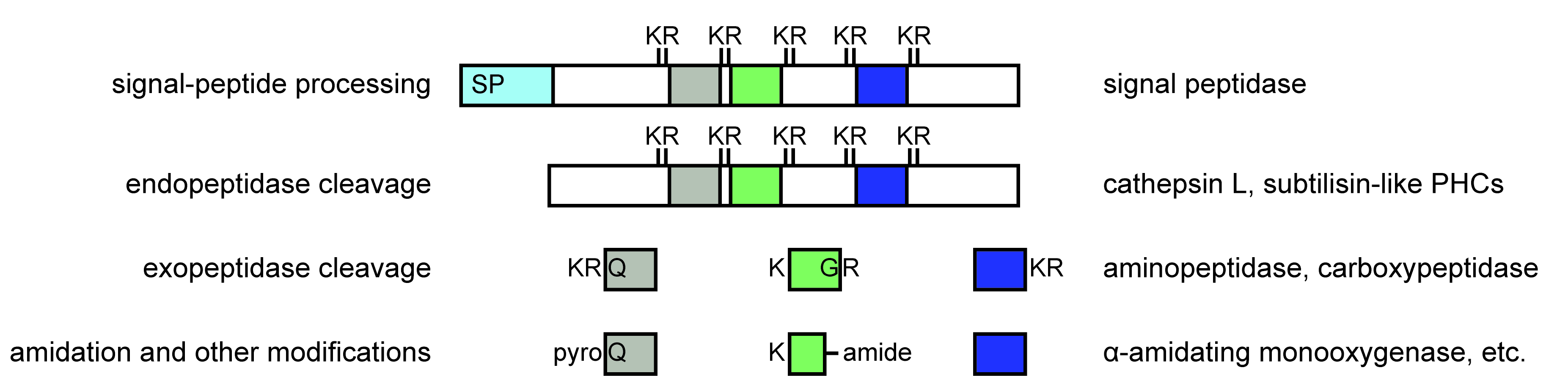
Neuropeptides are produced from precursor peptides
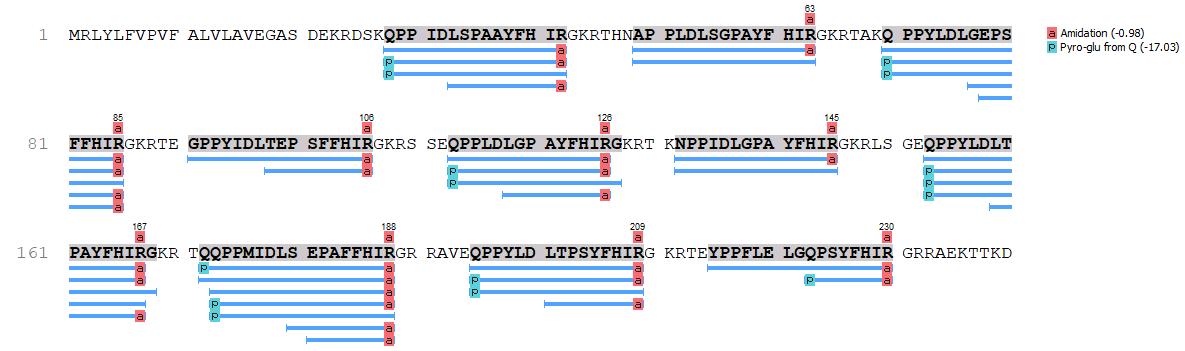
- mass spectrometry (LC-MS/MS) can identify the processed neuropeptides
- MetOH/acetic acid extraction of small peptides
- search also for modifications
Global view of the evolution of neuropeptides
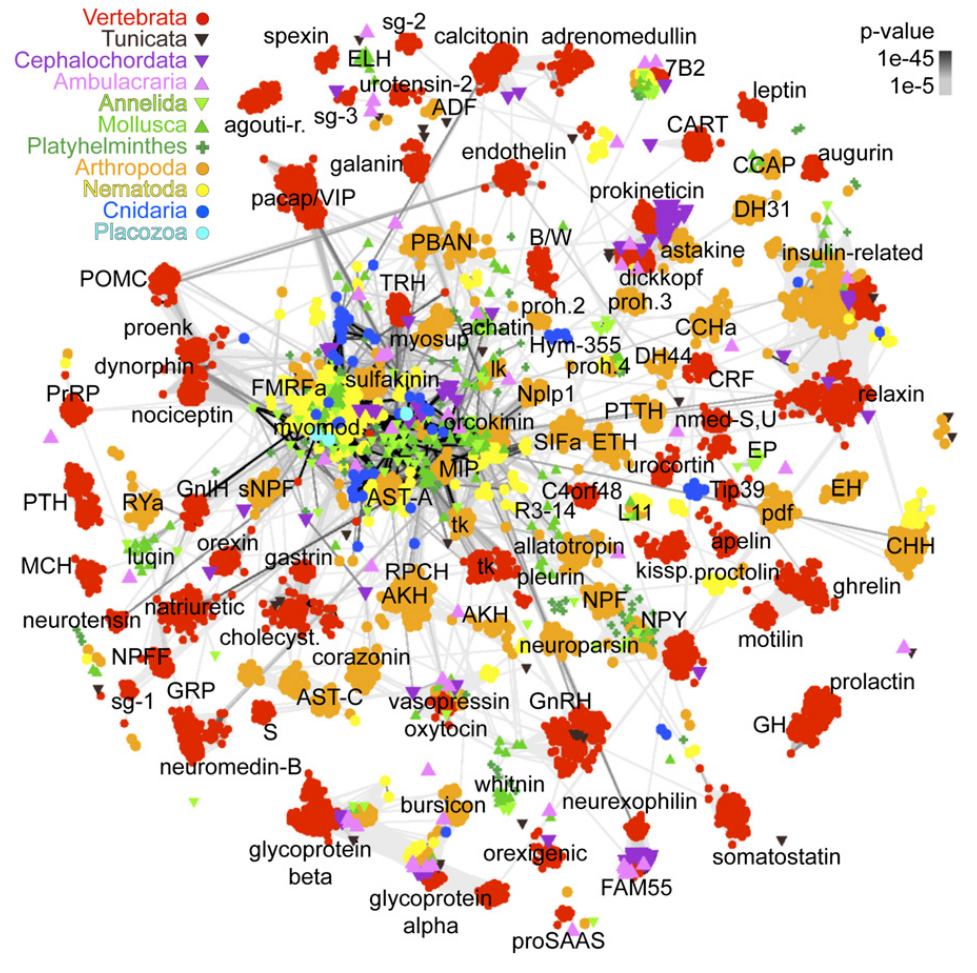
Global view of the evolution of neuropeptides
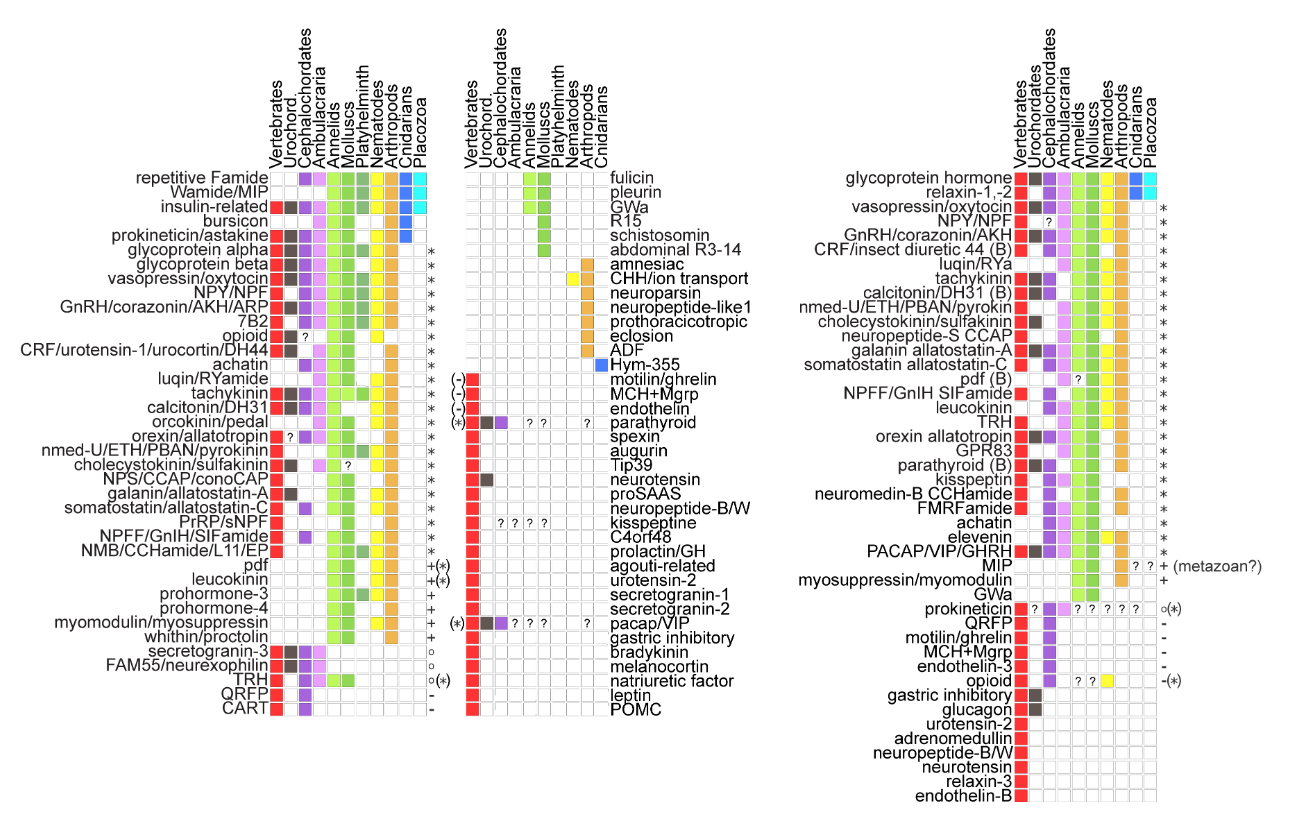
Tree of main animal lineages
Emerging marine model organisms


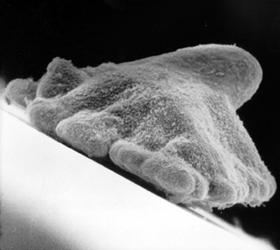
Image: Patrick Steinmetz
Nematostella vectensis
Platynereis dumerilii
Trichoplax adhaerens
Placozoa – no synapses, many peptides

Trichoplax adhaerens
- placozoans - simplest animals
- no neurons, no muscles
- upper and lower ciliated epithelium
- many neuropeptide-like molecules
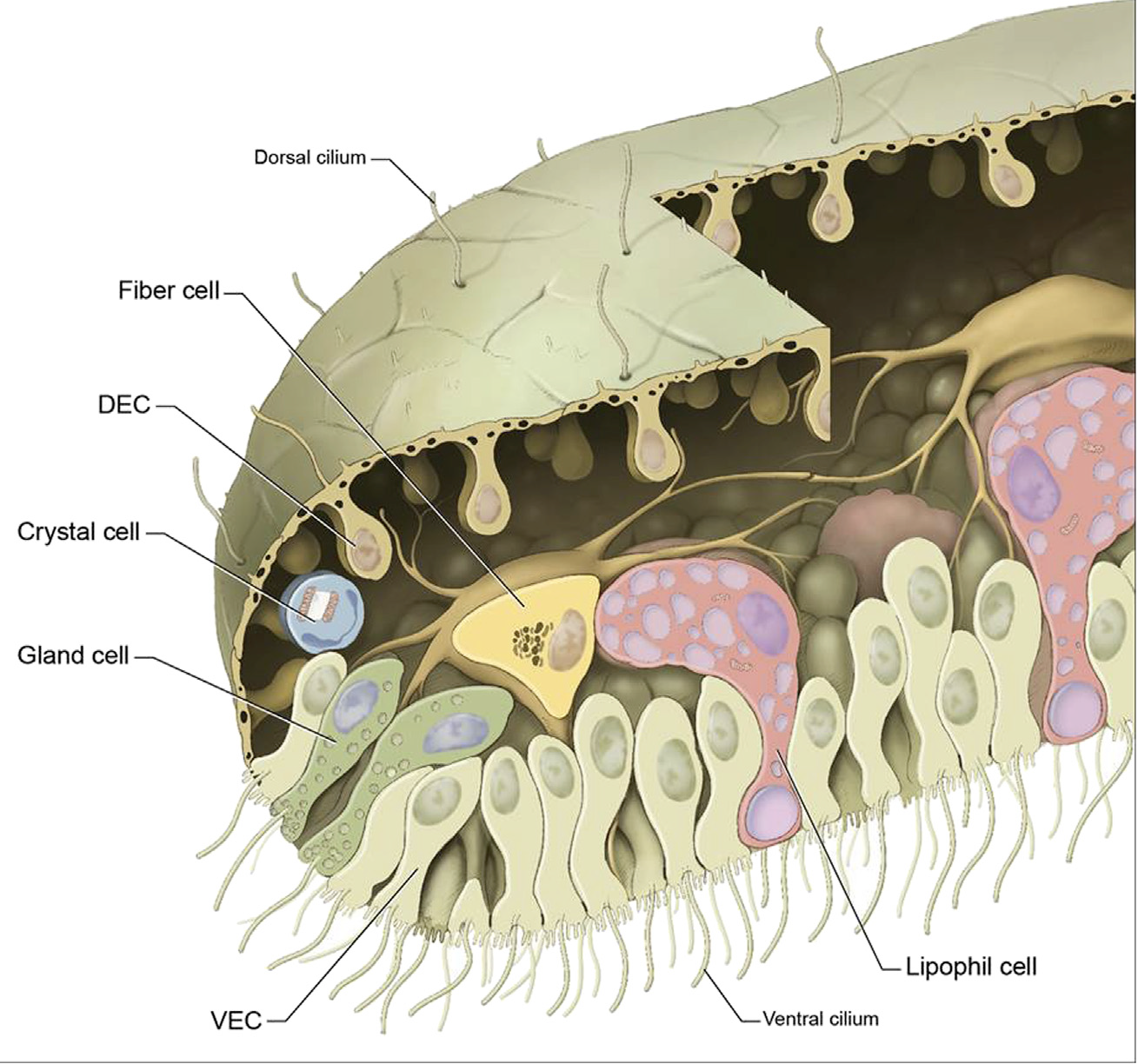
- few morphological cell types
Placozoa – fission
Placozoa – external digestion
- adhearance to substrate
- secretion of digestive enzymes
- external digestion
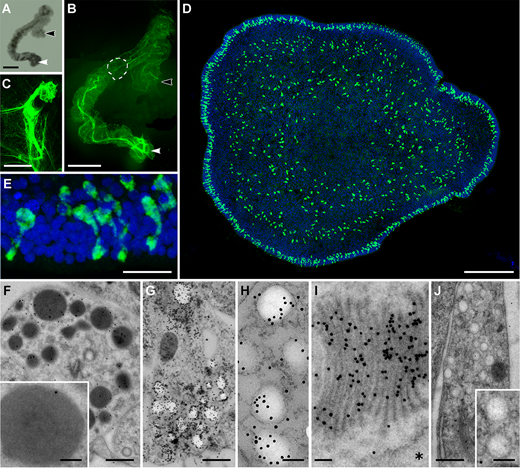
Placozoa – no synapses, many peptides
- neuropeptides are expressed in specific cell populations
- non-overlapping expression
- over 30 proneuropeptides
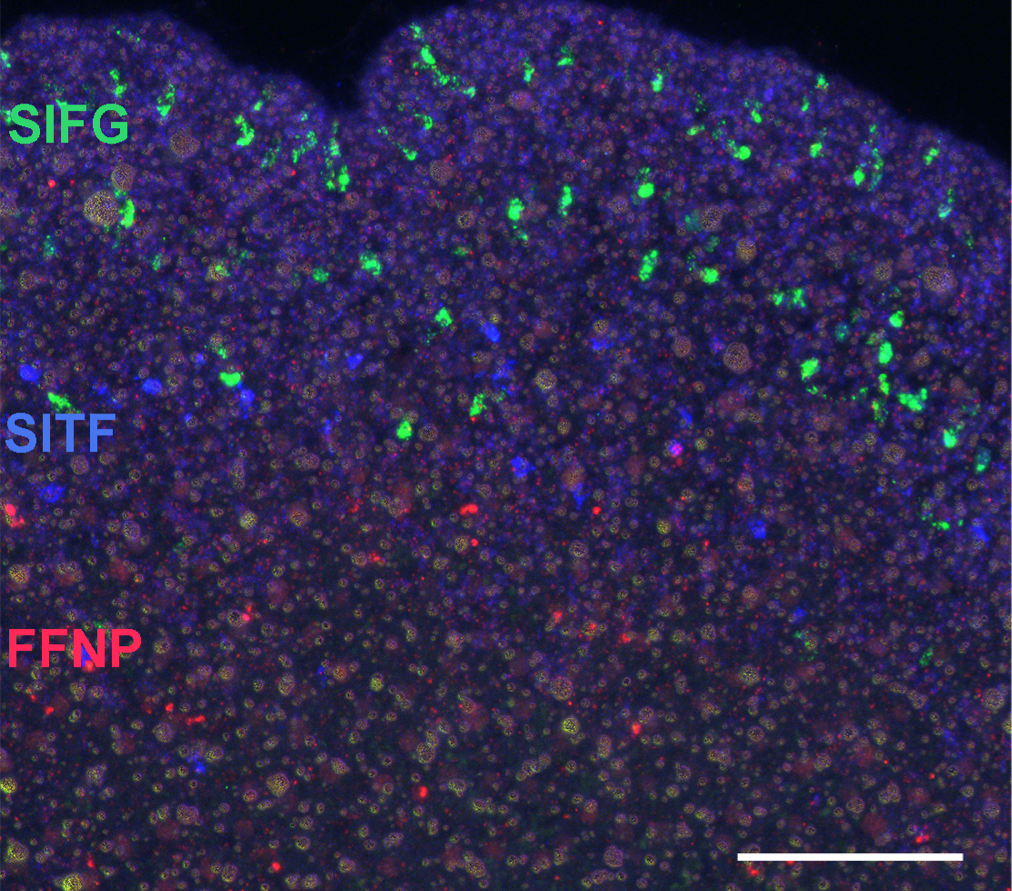
- peptidergic cells tile the epithelium
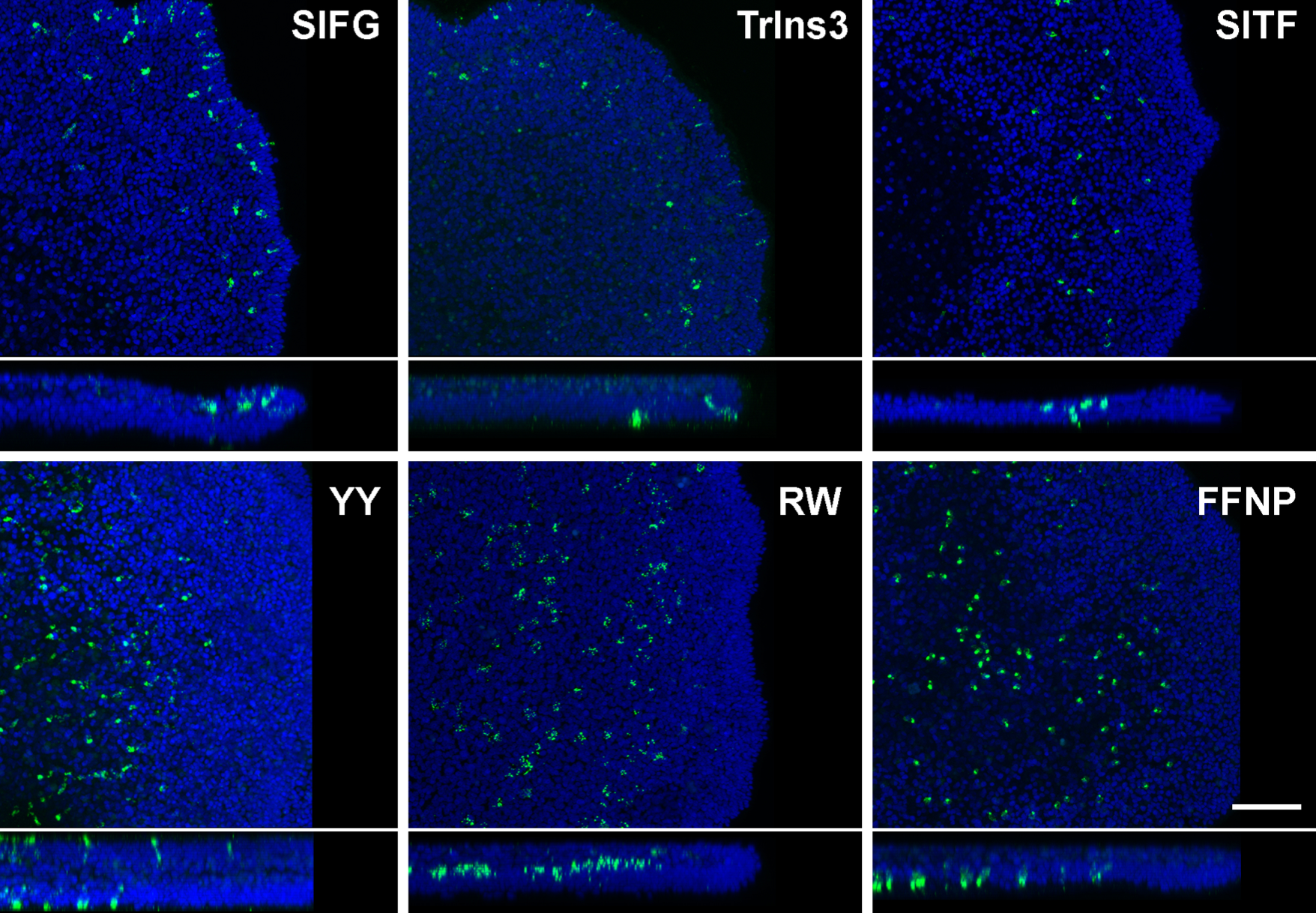
Placozoa – peptide effects

- neuropeptides influence placozoan behaviour
LF peptide
Placozoa – peptide effects
SIFGamide peptide
WPPF peptide
How can we identify and characterise neuropeptide receptors?
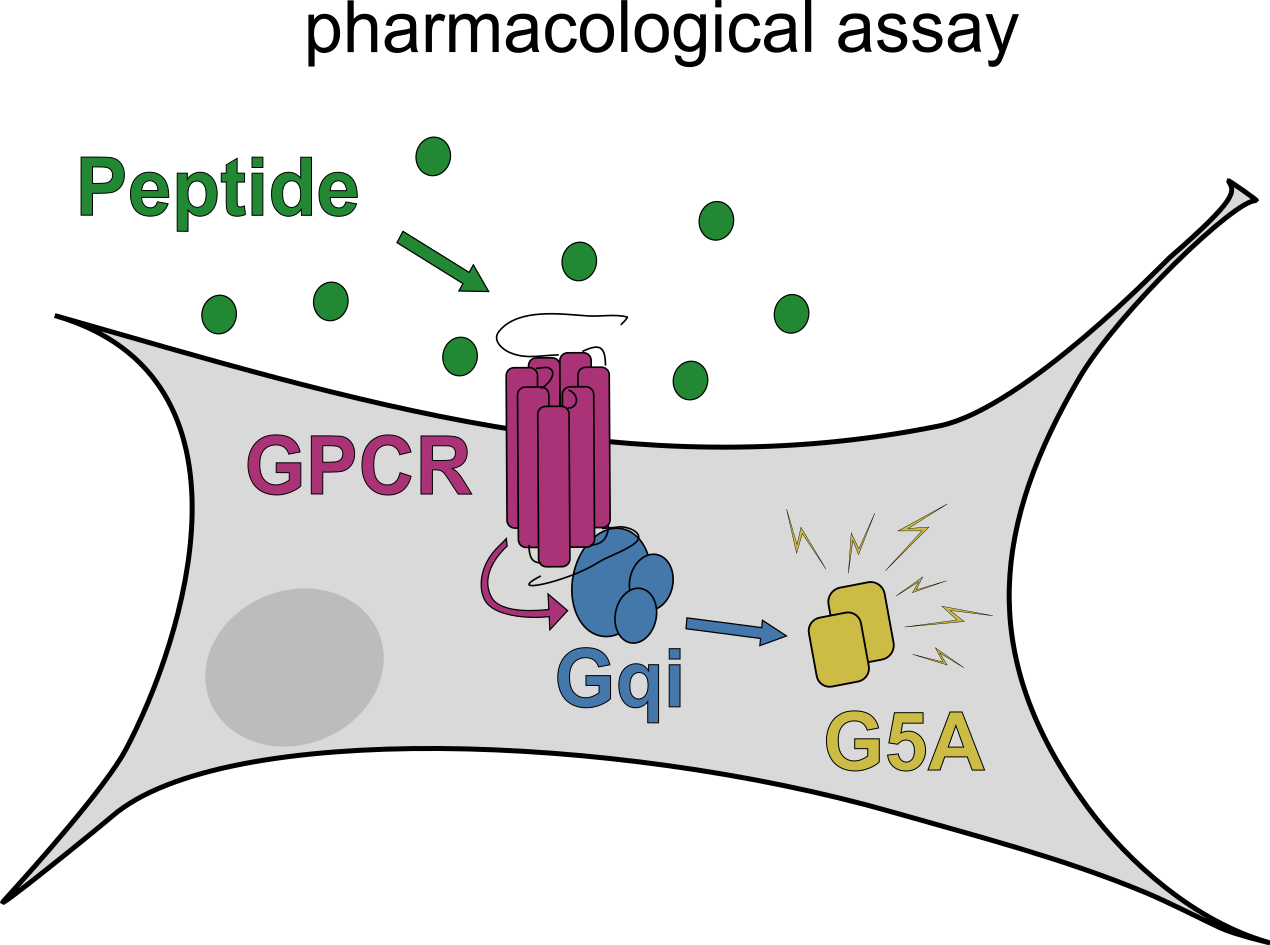
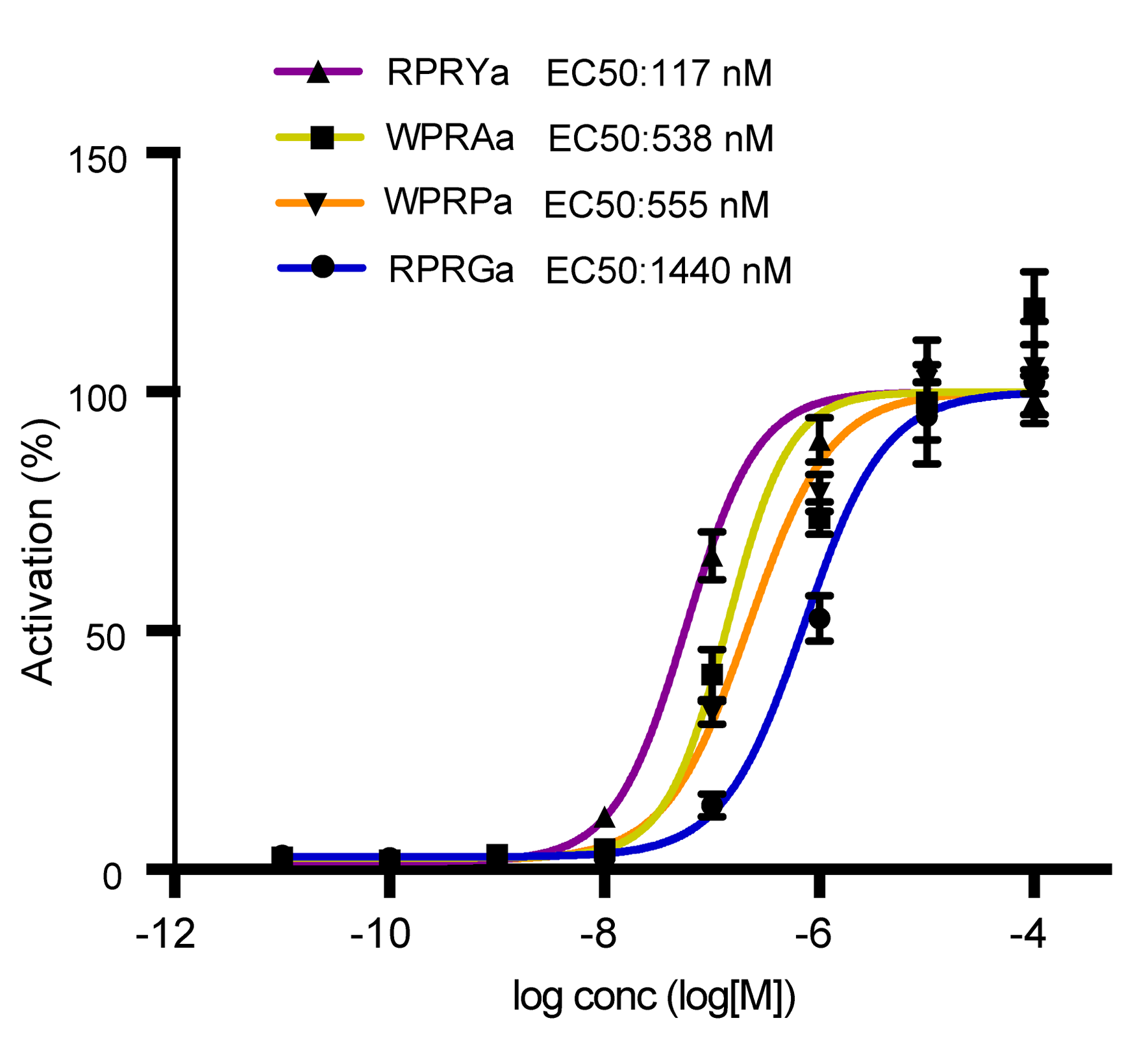
Identification of placozoan neuropeptide GPCRs
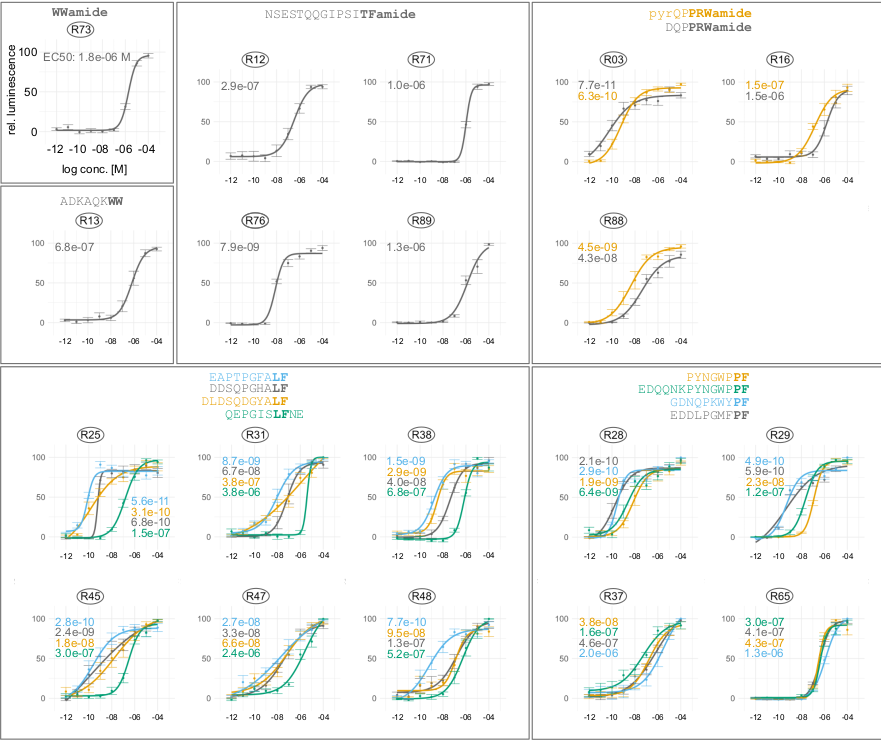
Identification of placozoan neuropeptide GPCRs
Neuropeptides in Cnidaria
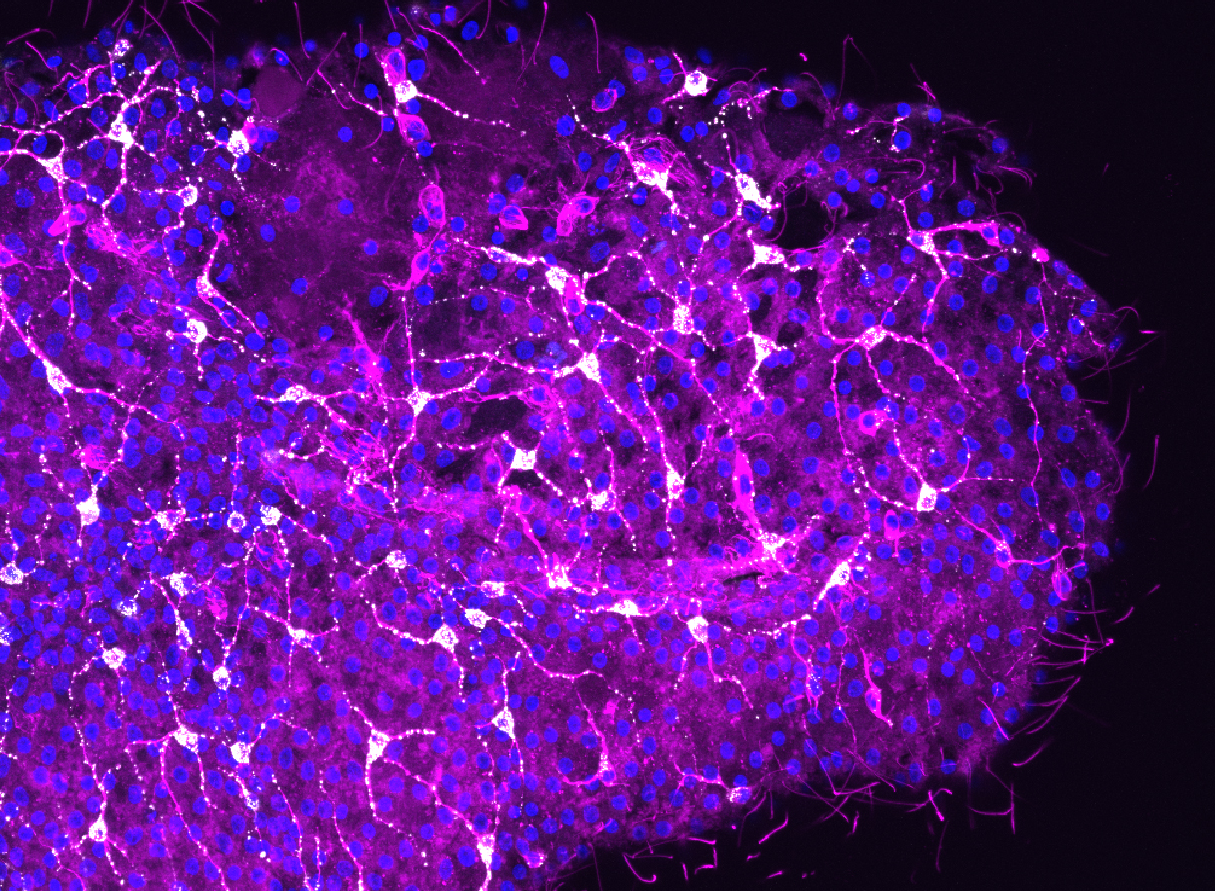
Peptidergic nerve nets
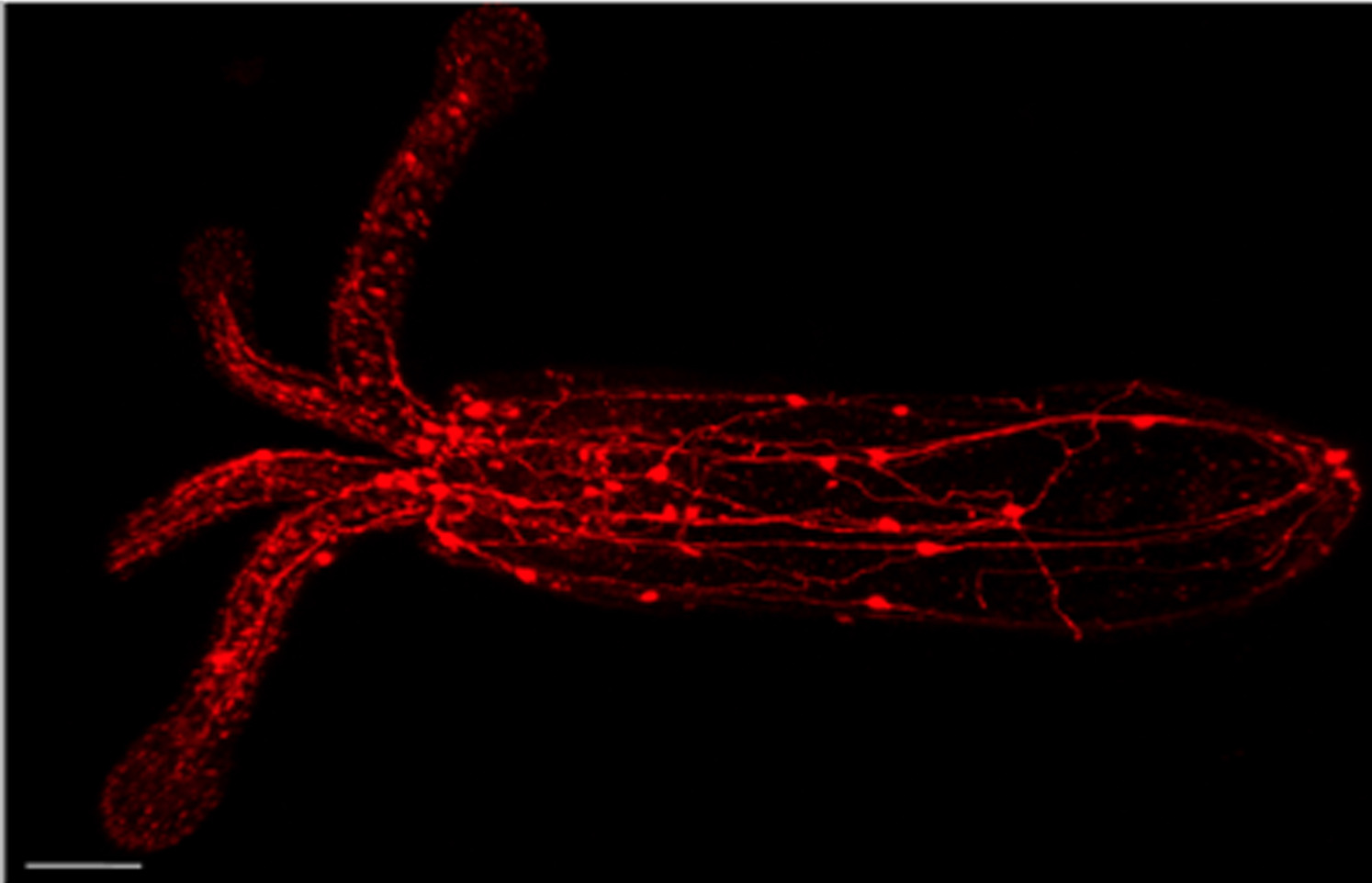
LWamide transgene
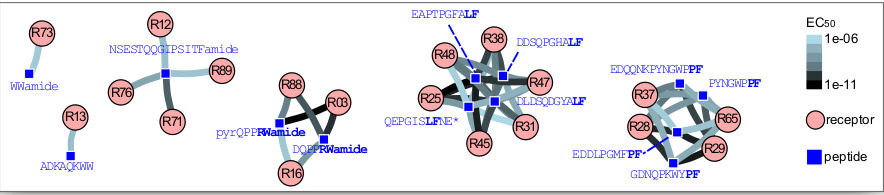
Large-scale GPCR-peptide screen in Nematostella

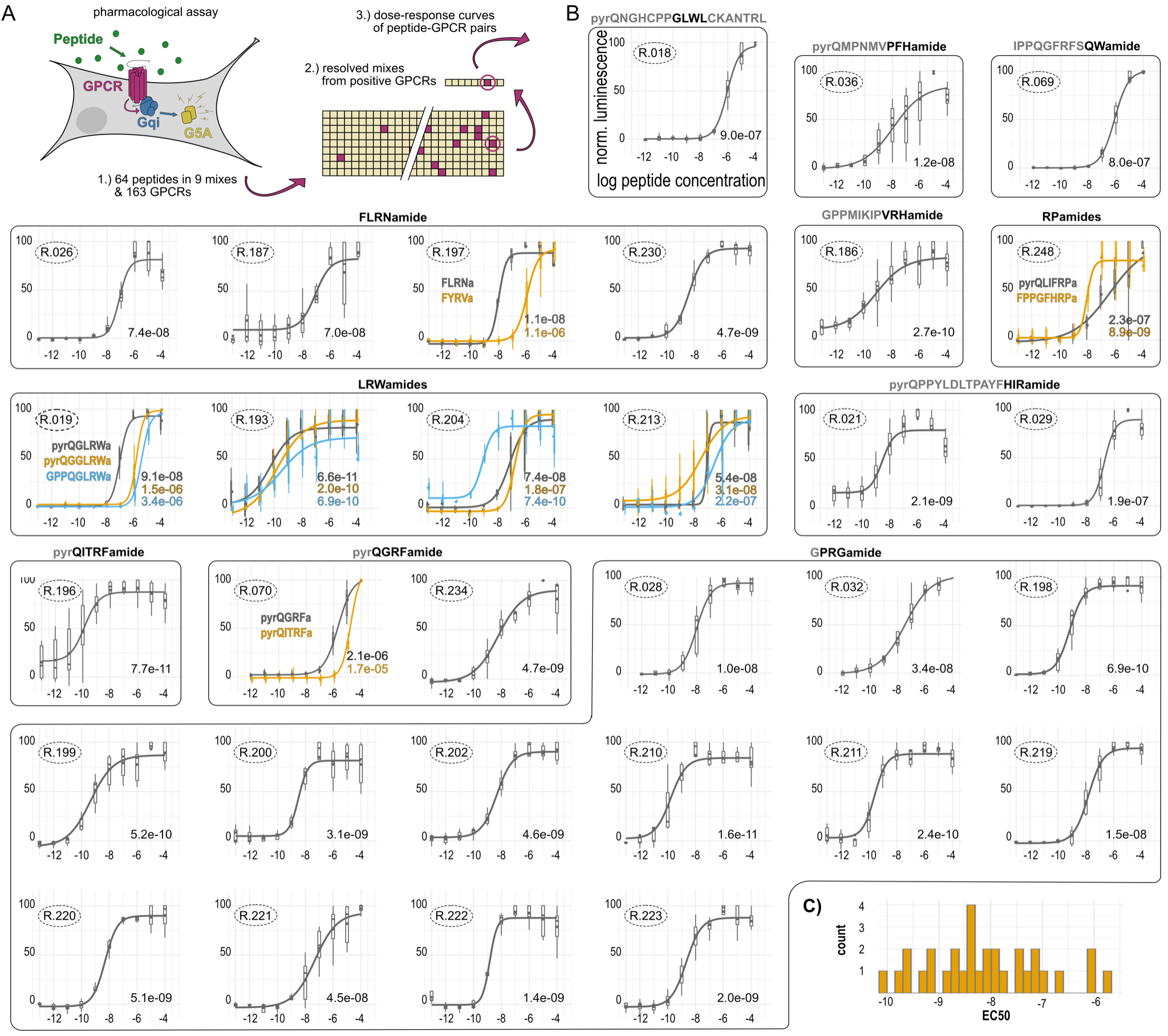
Mapping to single-cell data
A peptidergic connectome
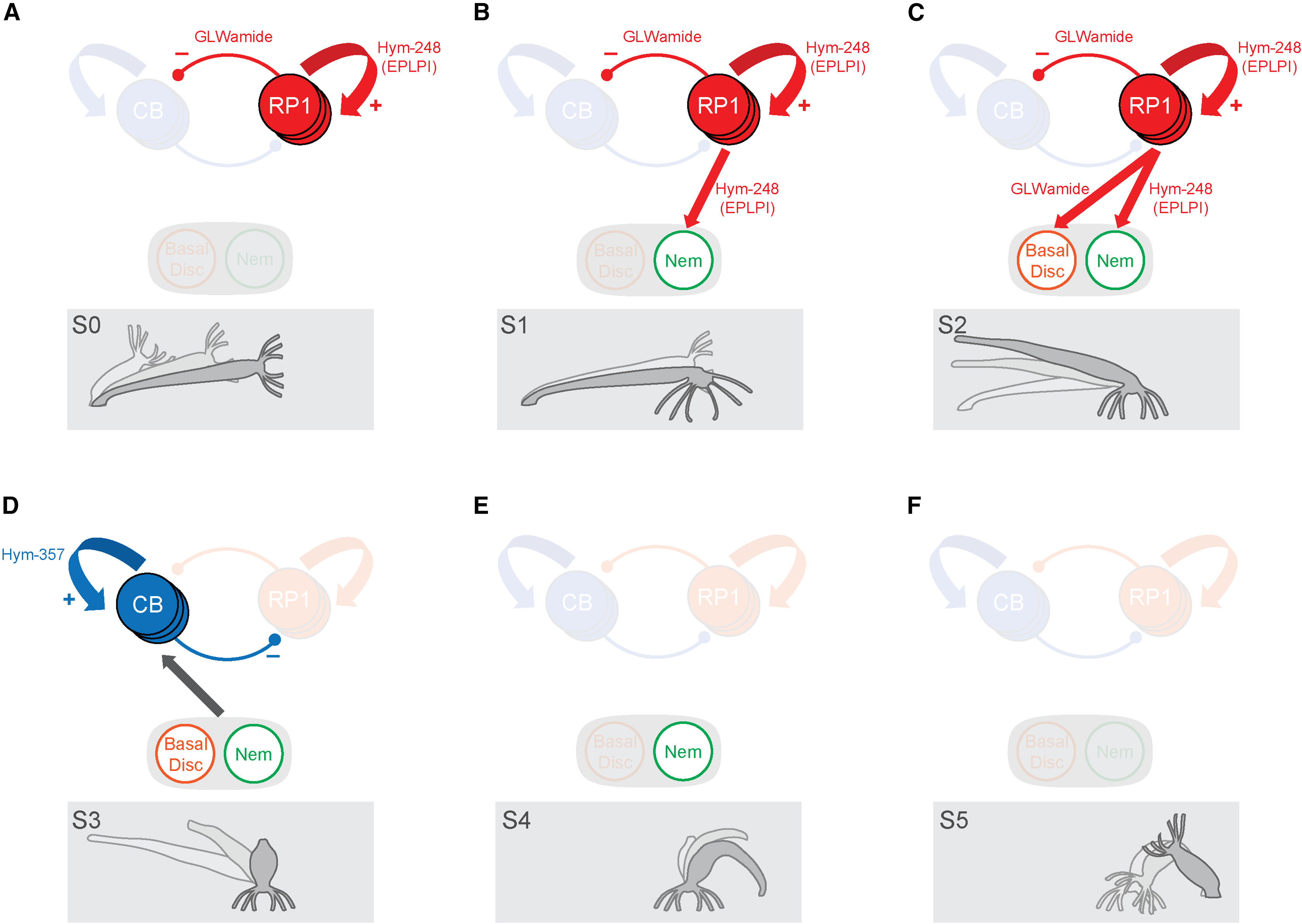
Neuropeptides can induce behaviour in cnidaria
Hydra vulgaris
Hym-248 neuropeptide induces somersaulting
A peptidergic model for Hydra somersaulting
Peptide-controlled spawning in Clytia
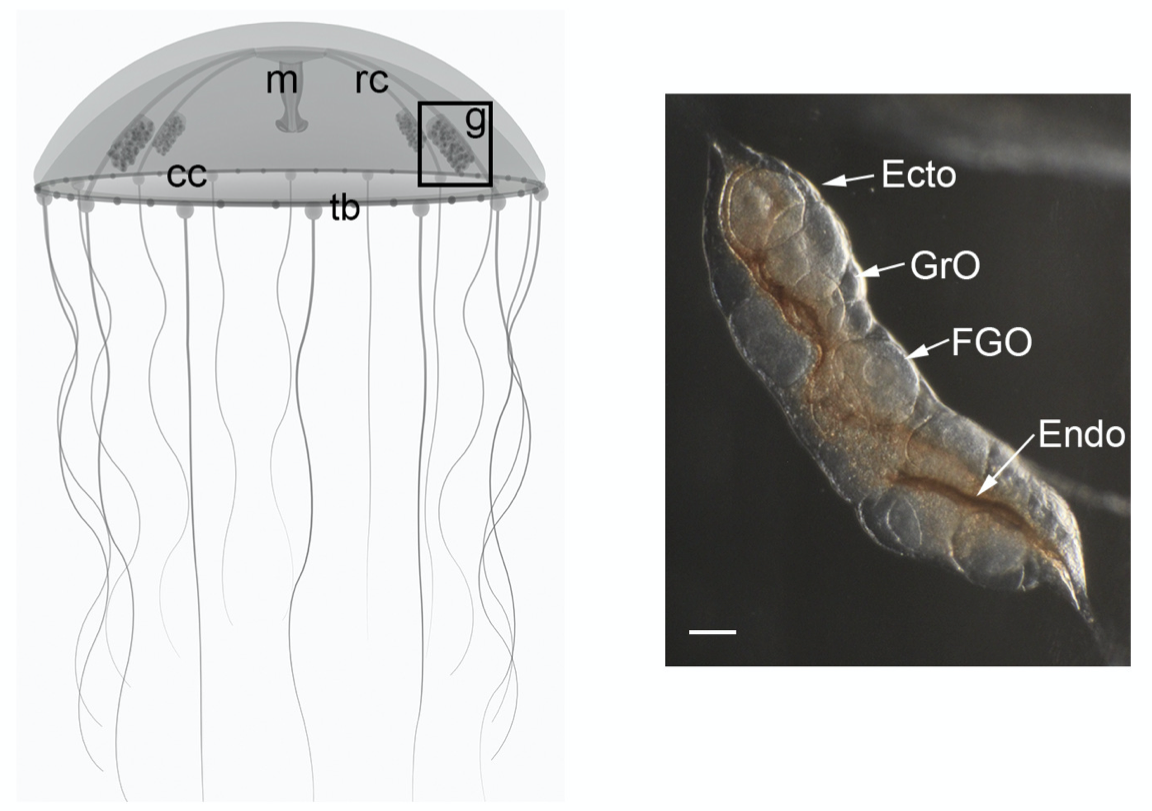
Peptide-controlled spawning in Clytia
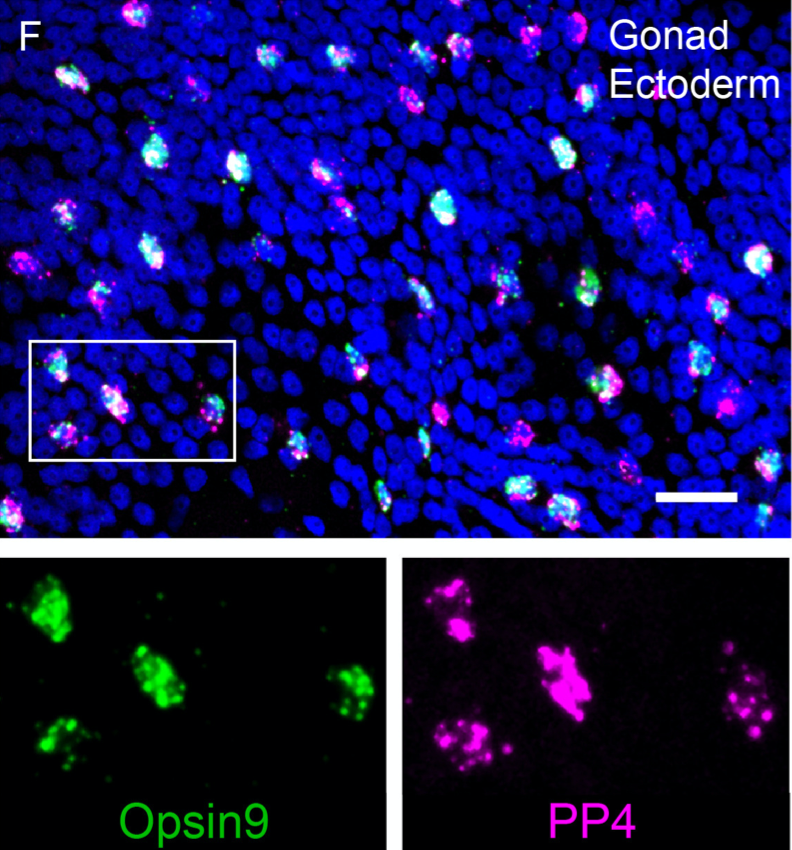
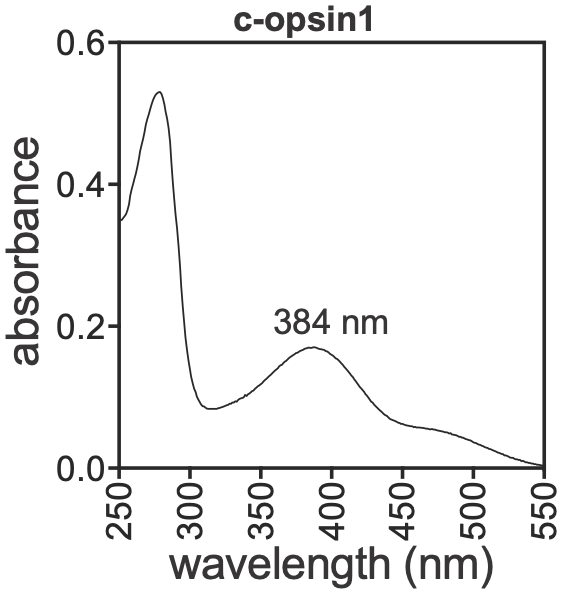
- an opsin coexpresses with the maturation-inducing neuropeptide
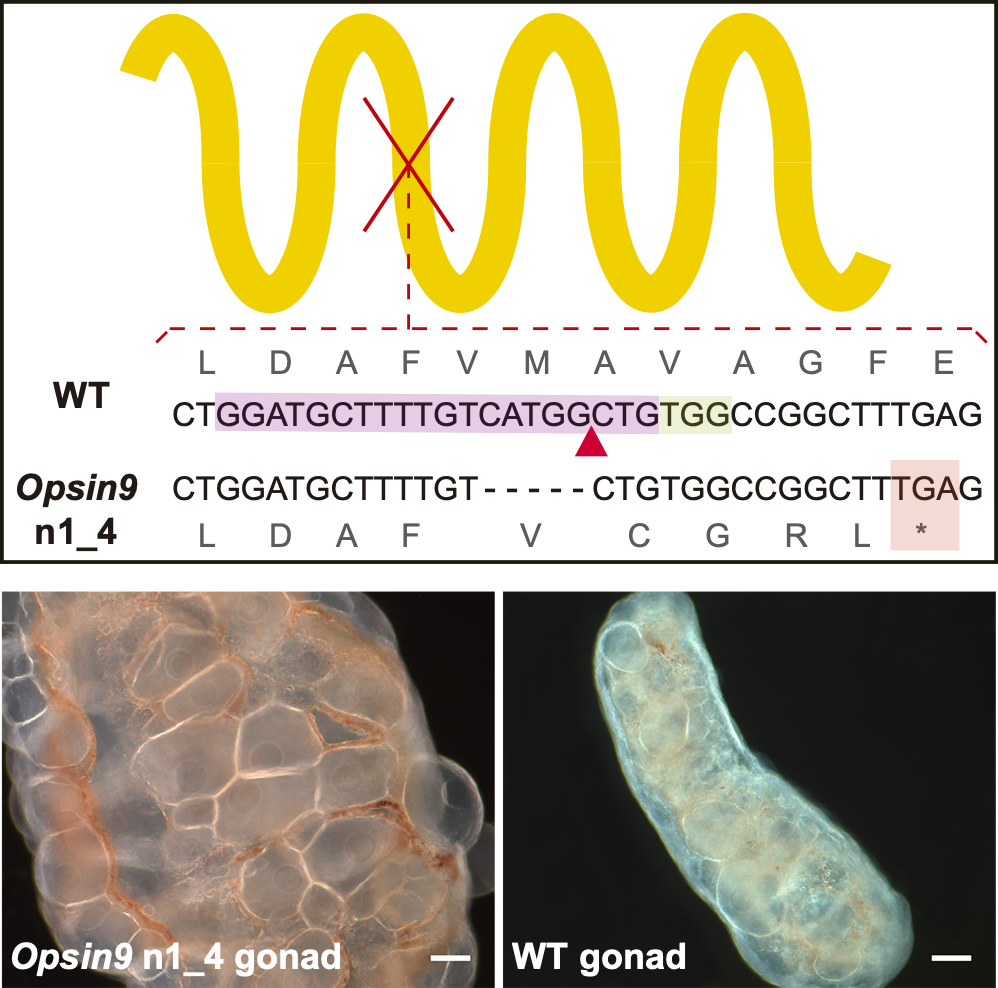
- CRISPR knockout animals for the opsin do not spawn in light
Peptide-controlled spawning in Clytia
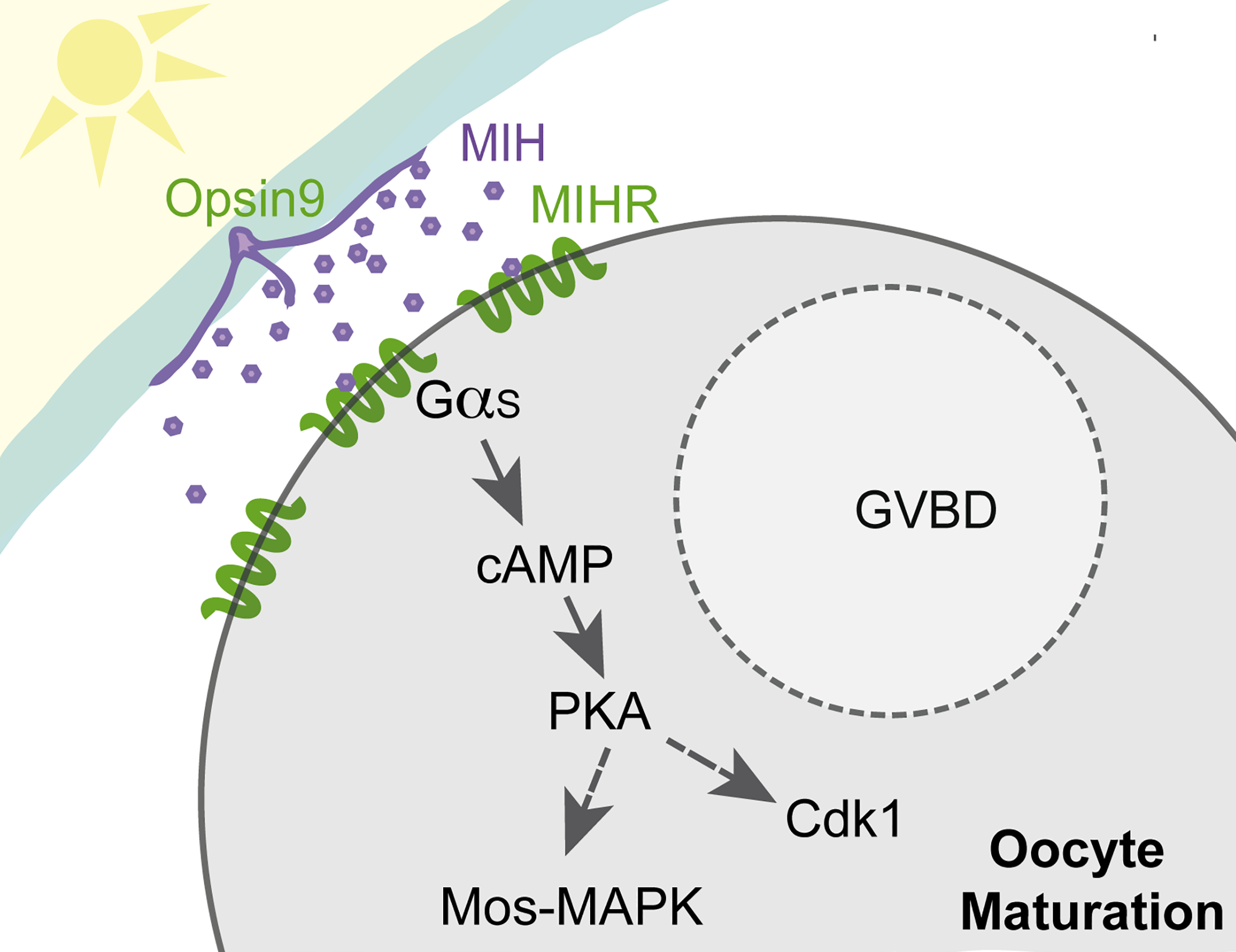
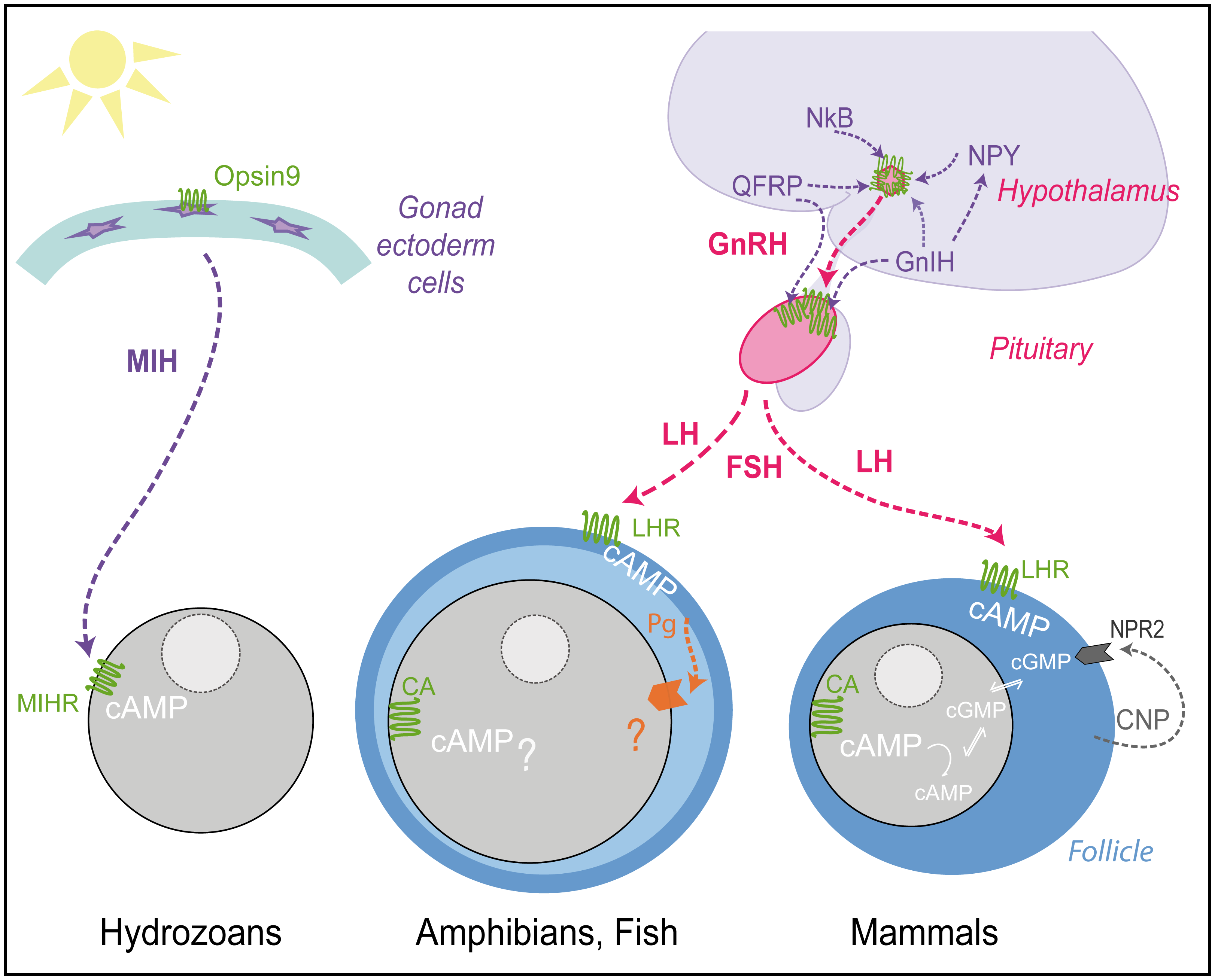
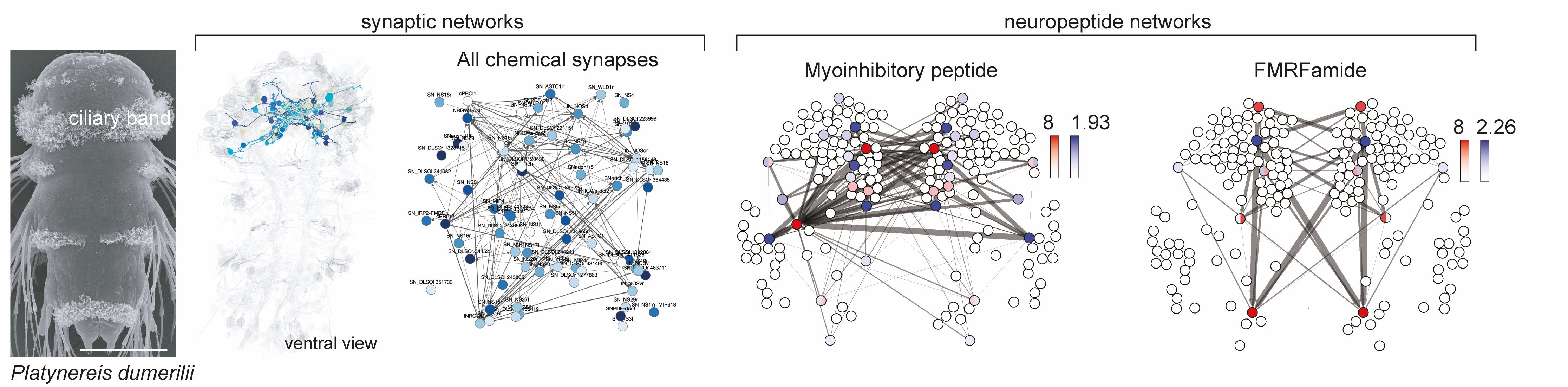
The connectome is multilayered
The connectome is multilayered
Platynereis dumerilii

- breeding culture, full life-cycle
- embryos daily, year round
- genome sequence
- microinjection, transgenesis
- neuron-specific promoters and antibodies
- knock-out lines
- neuronal connectome
- whole-body neuronal activity imaging
- whole-animal pharmacology by bath application 😎
Platynereis dumerilii
Spawning
movie by Albrecht Fischer
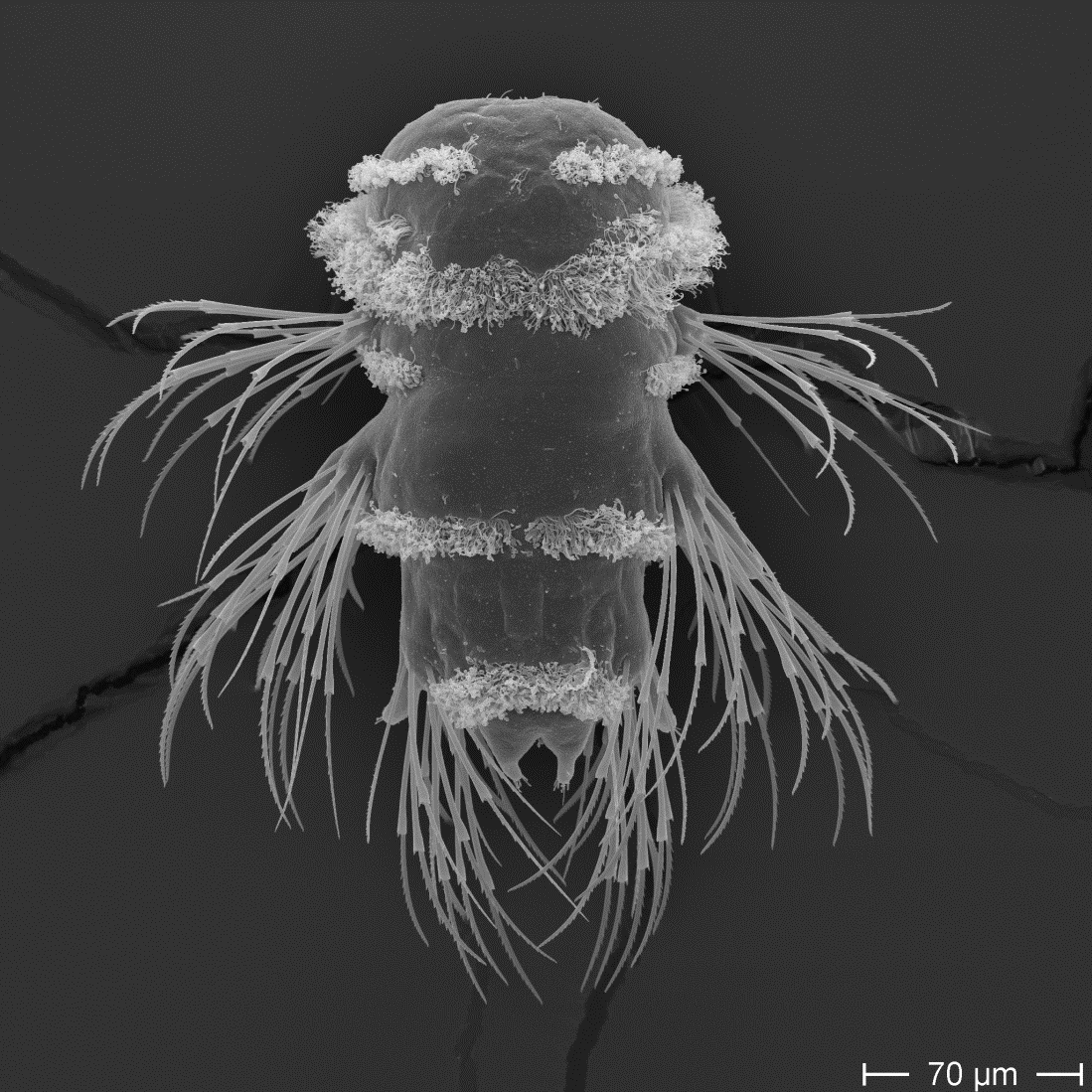
Synchronously developing larvae
Whole-body volume EM of an entire three-day-old larva
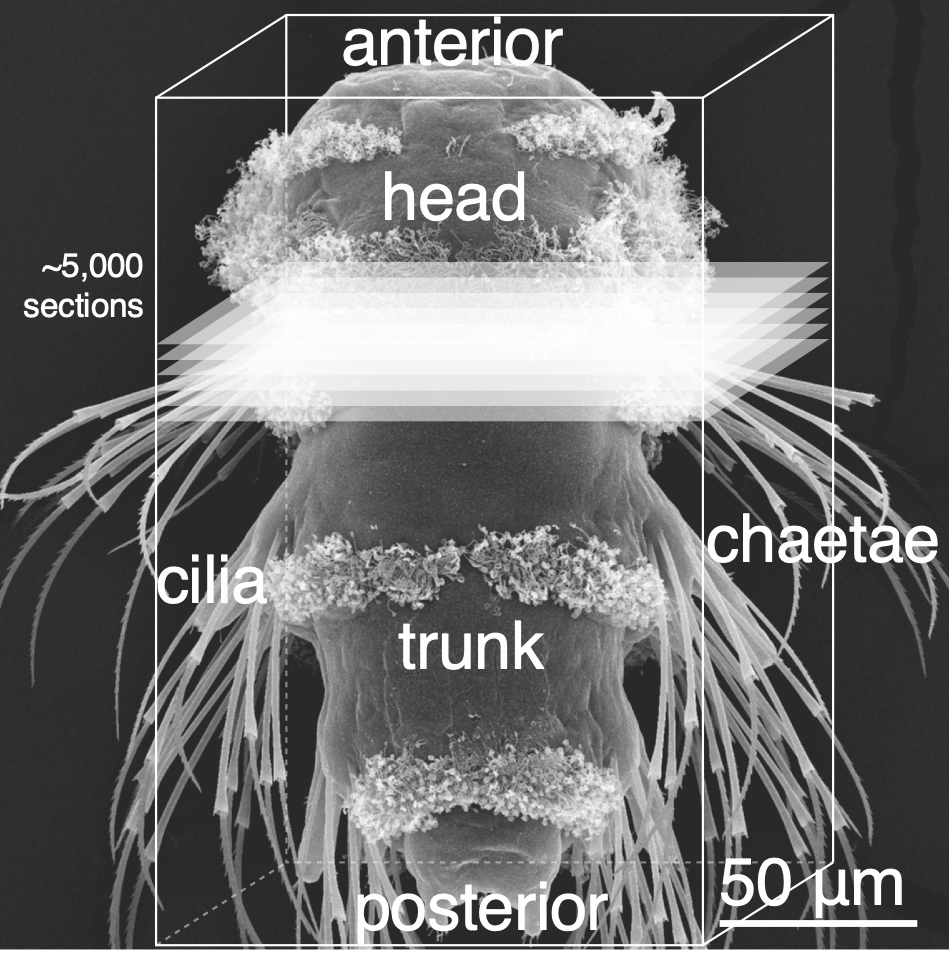
Whole-body connectome of a segmented annelid larva
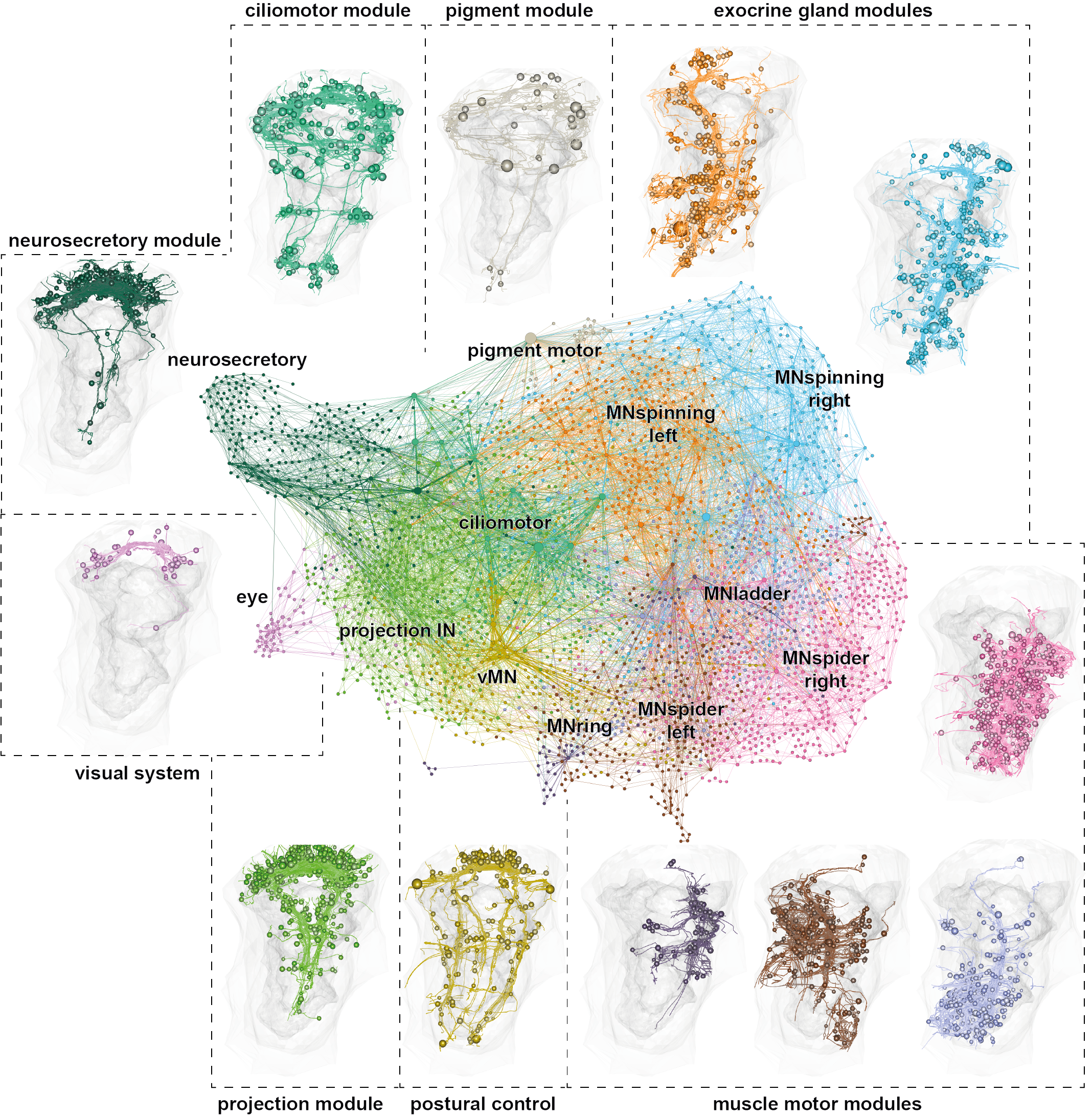
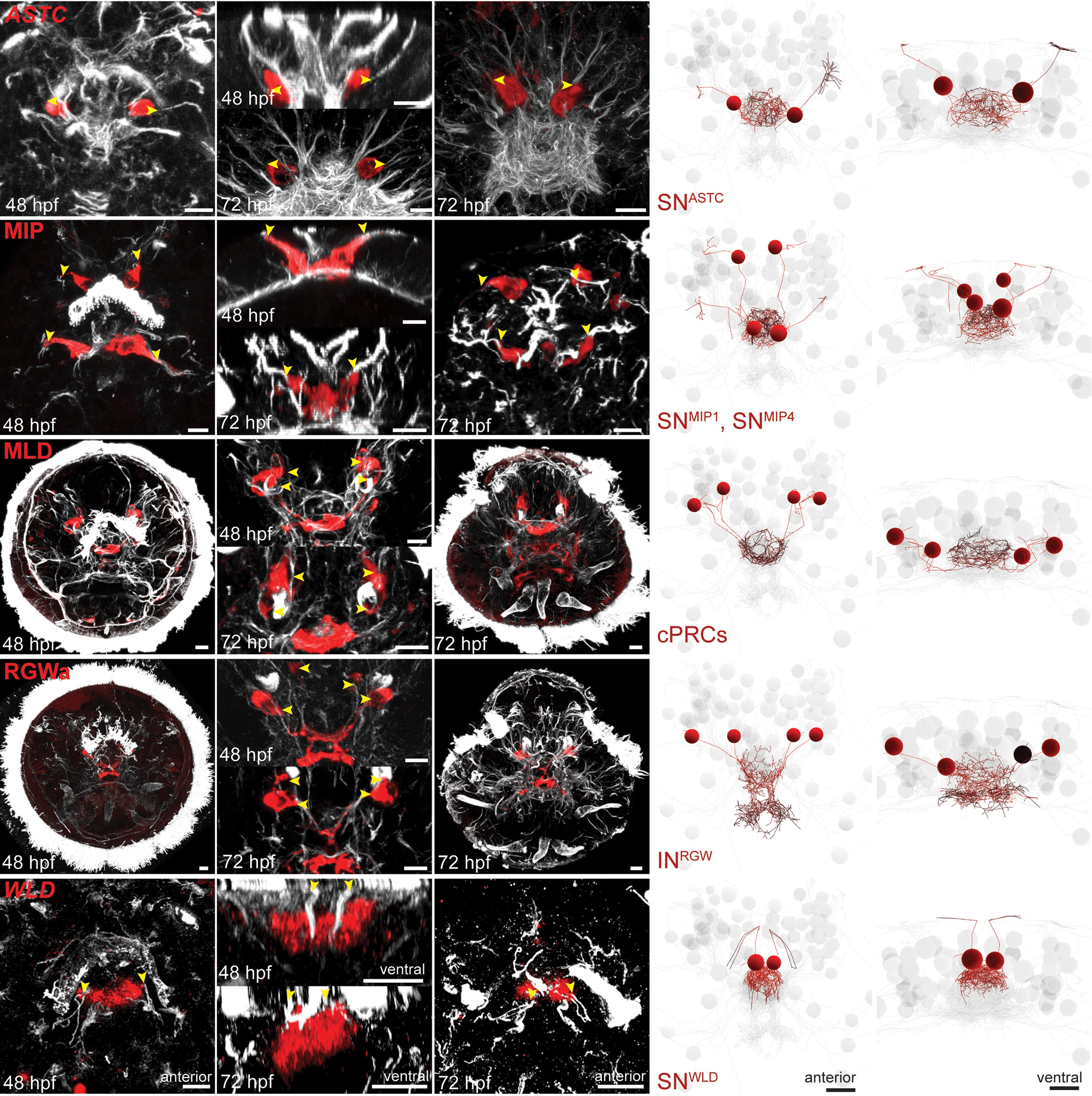
The nervous system of the larva
~2,000 neurons
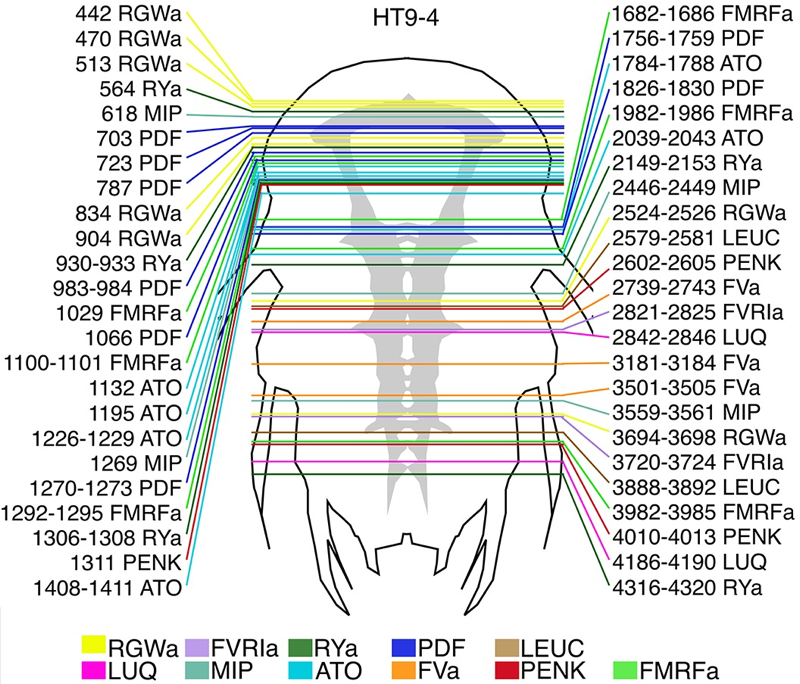
Synaptic connectome
Many peptides and modulators in the circuit
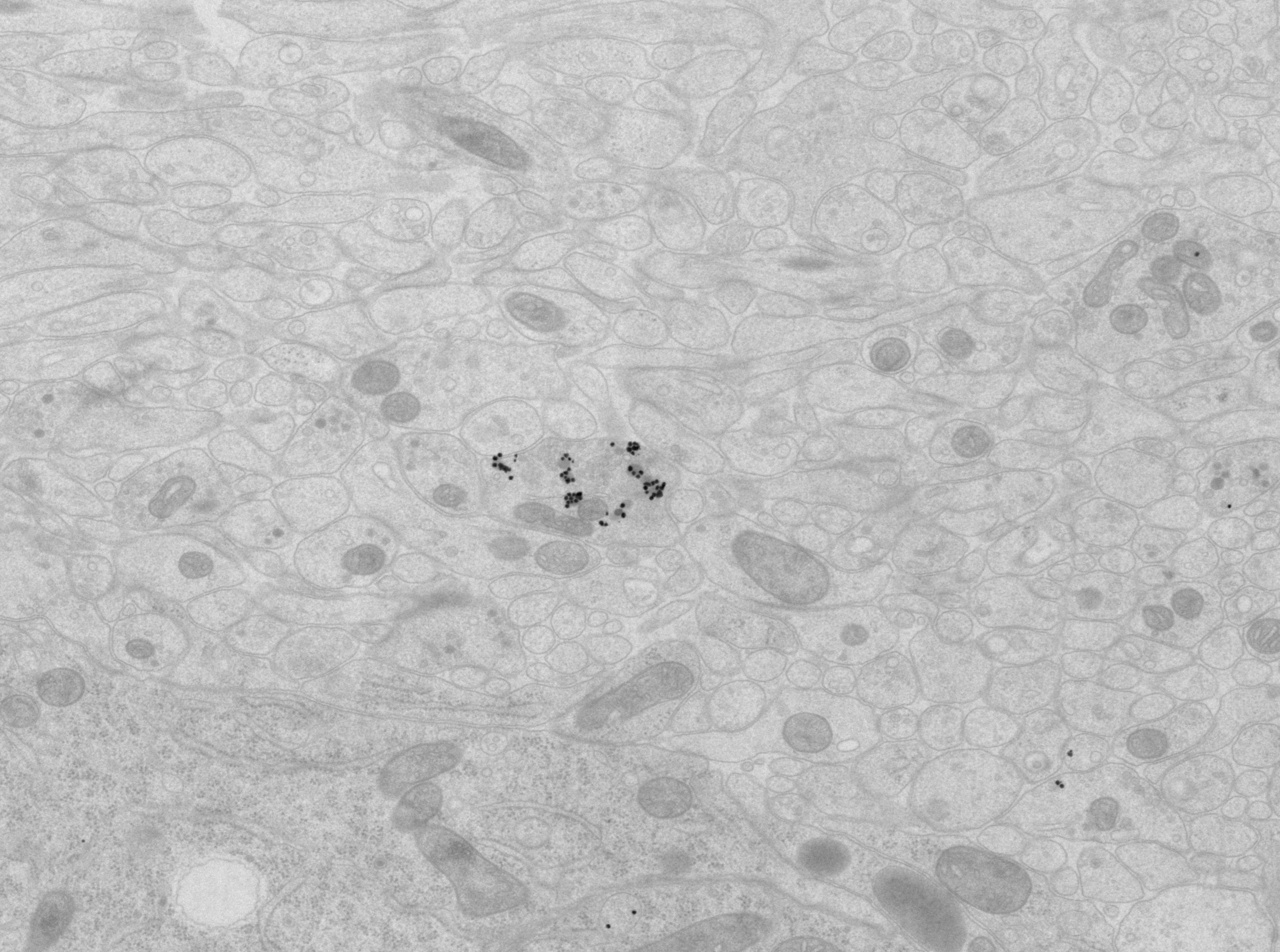
Multiplex immunogold for neuropeptides in the EM volume
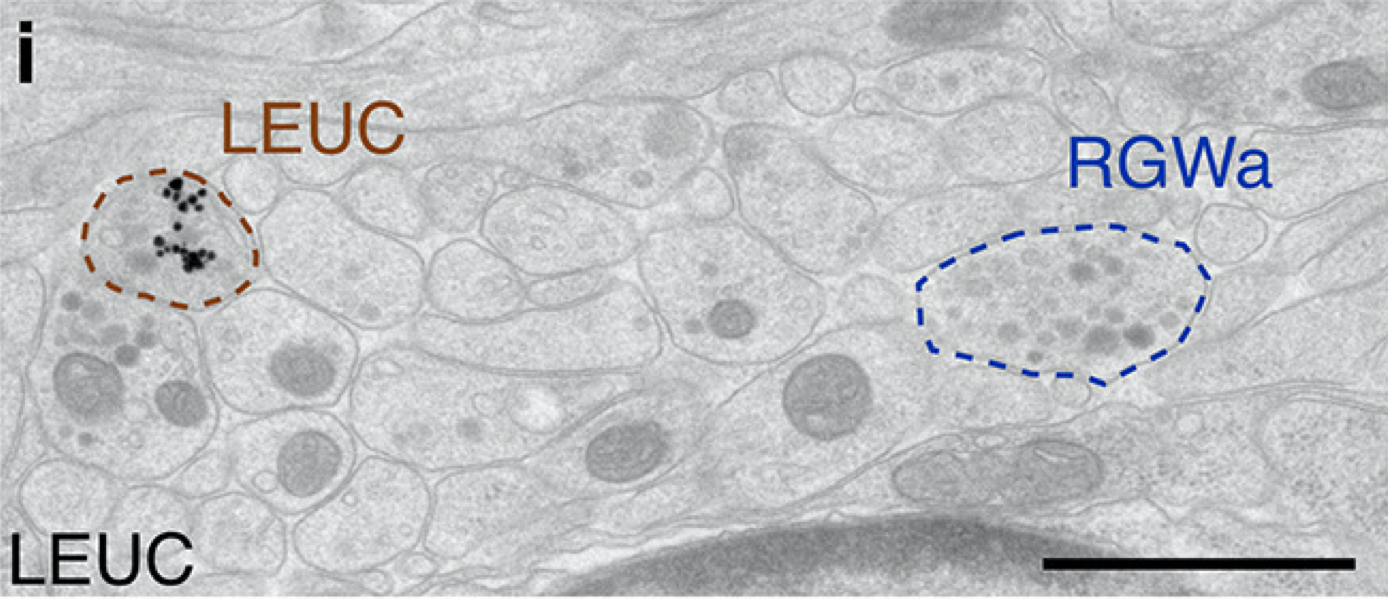
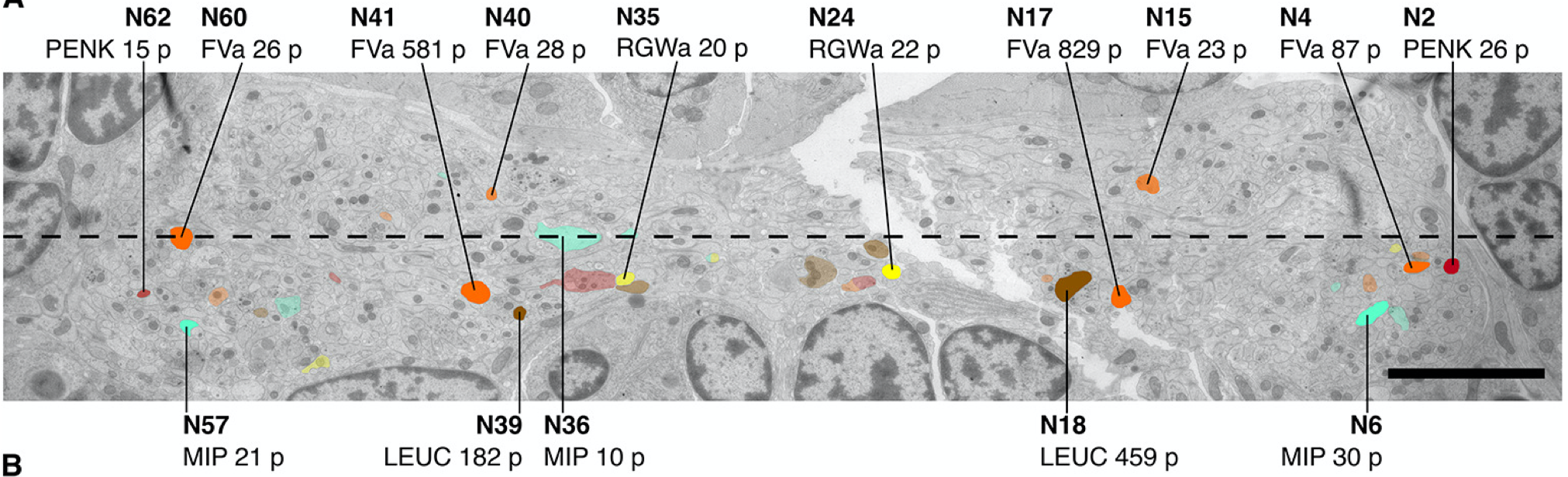
Mapping of (pro)neuropeptides

UV response in Platynereis larvae

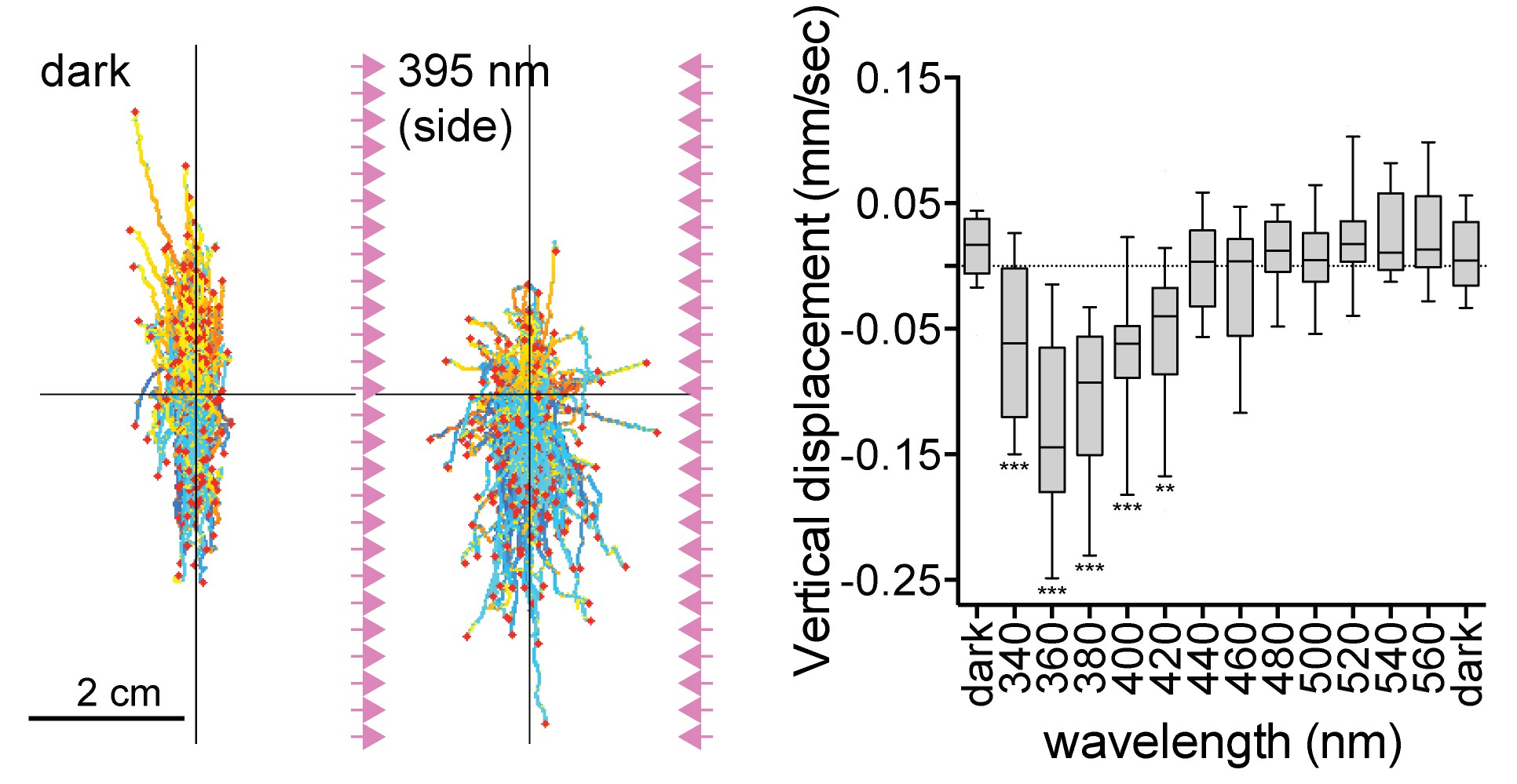
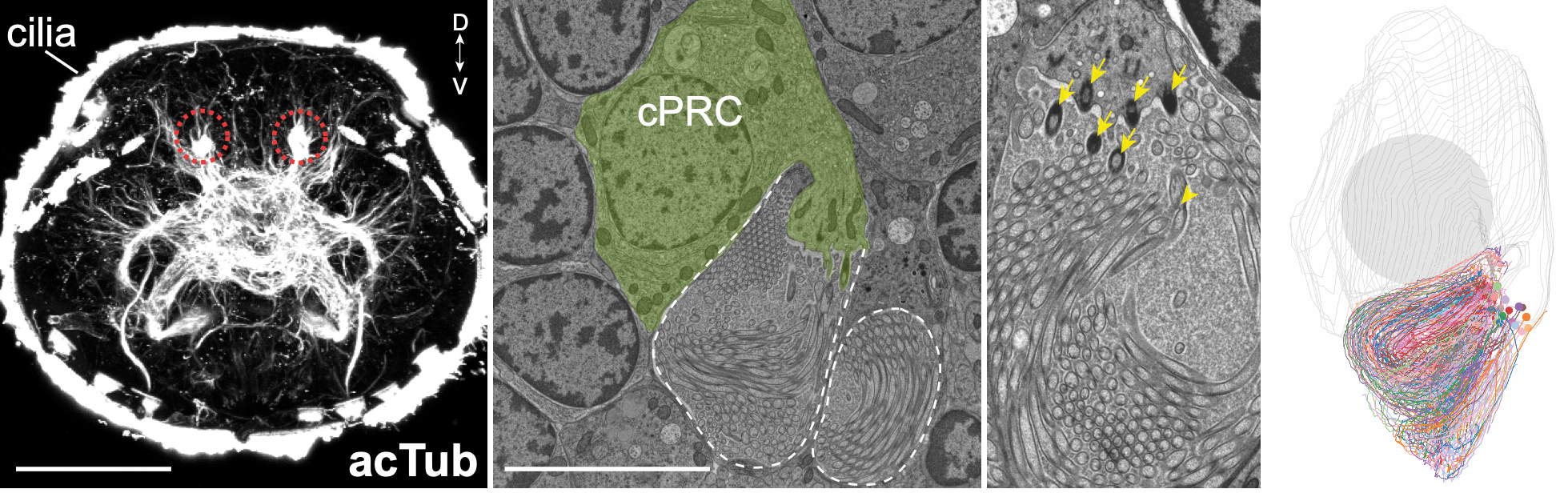
UV-responding brain ciliary photoreceptors (cPRCs)
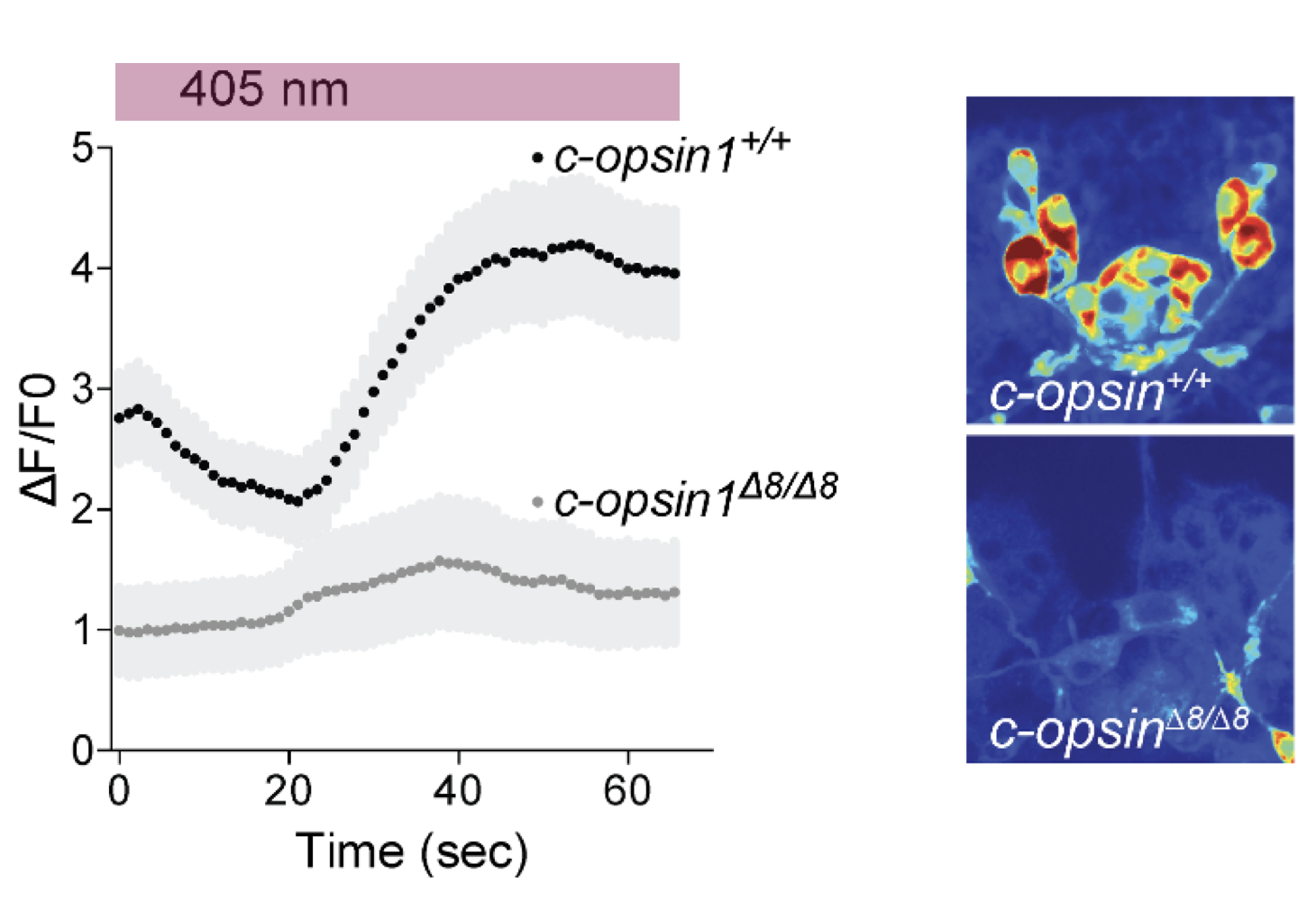
No UV avoidance in c-opsin1 knockouts
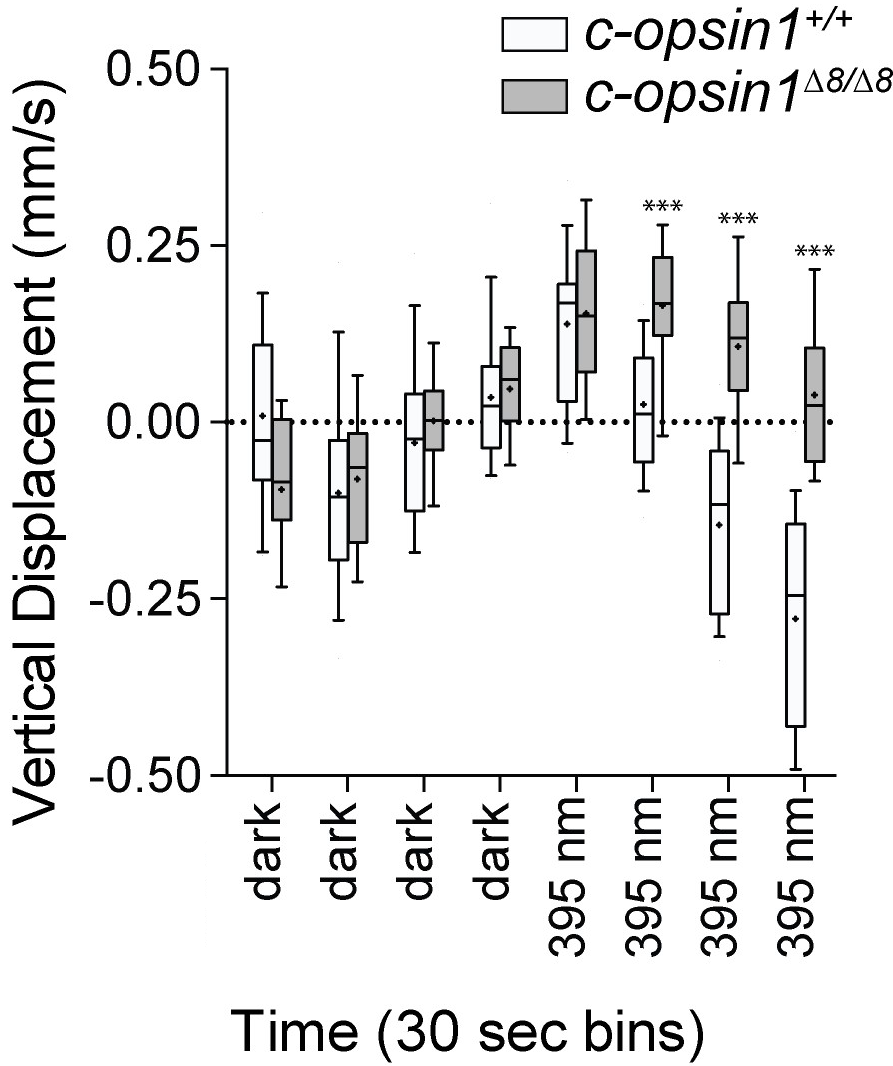
No cPRC response in c-opsin1 knockouts
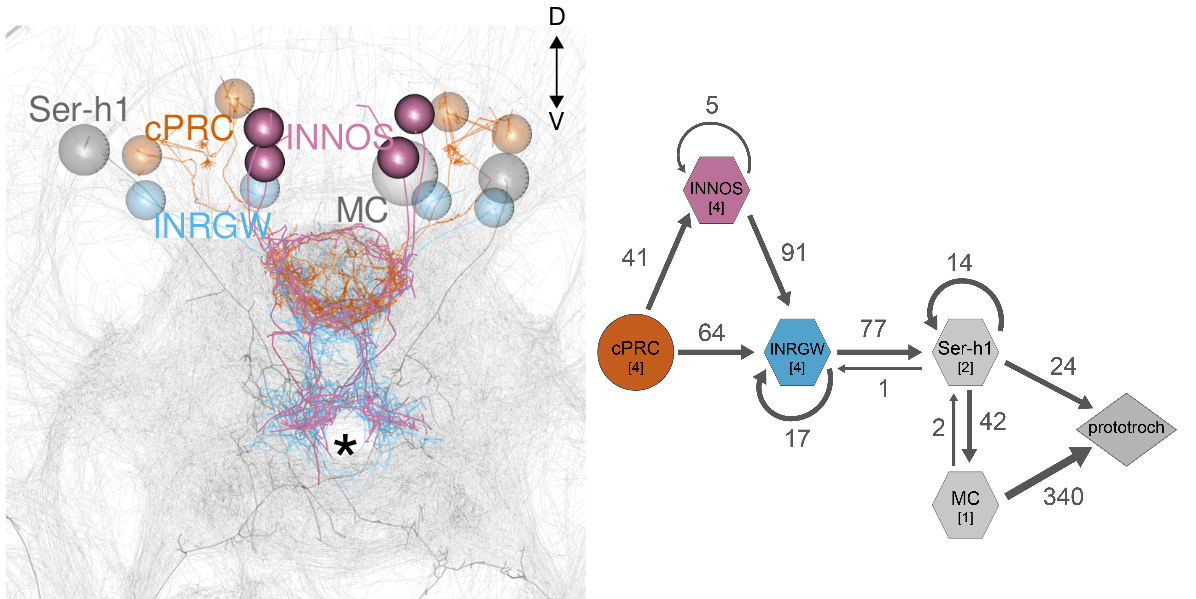
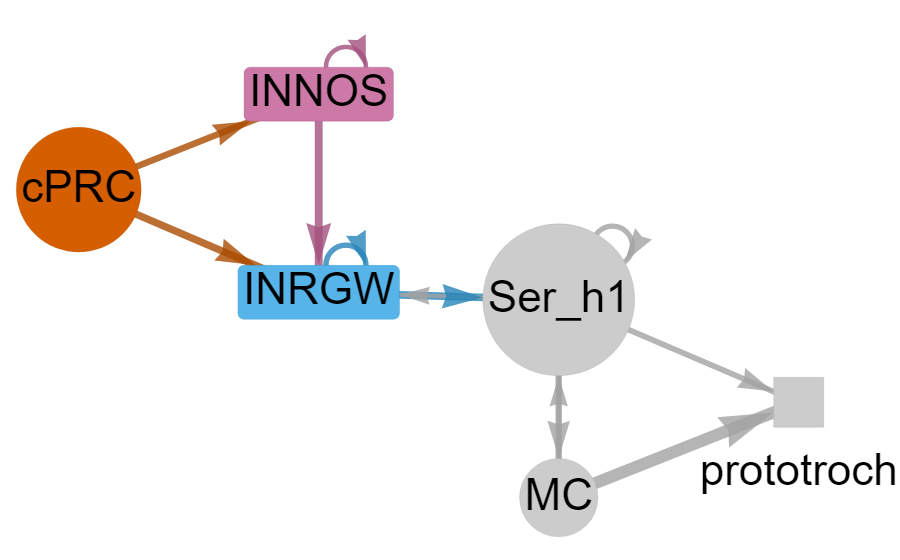
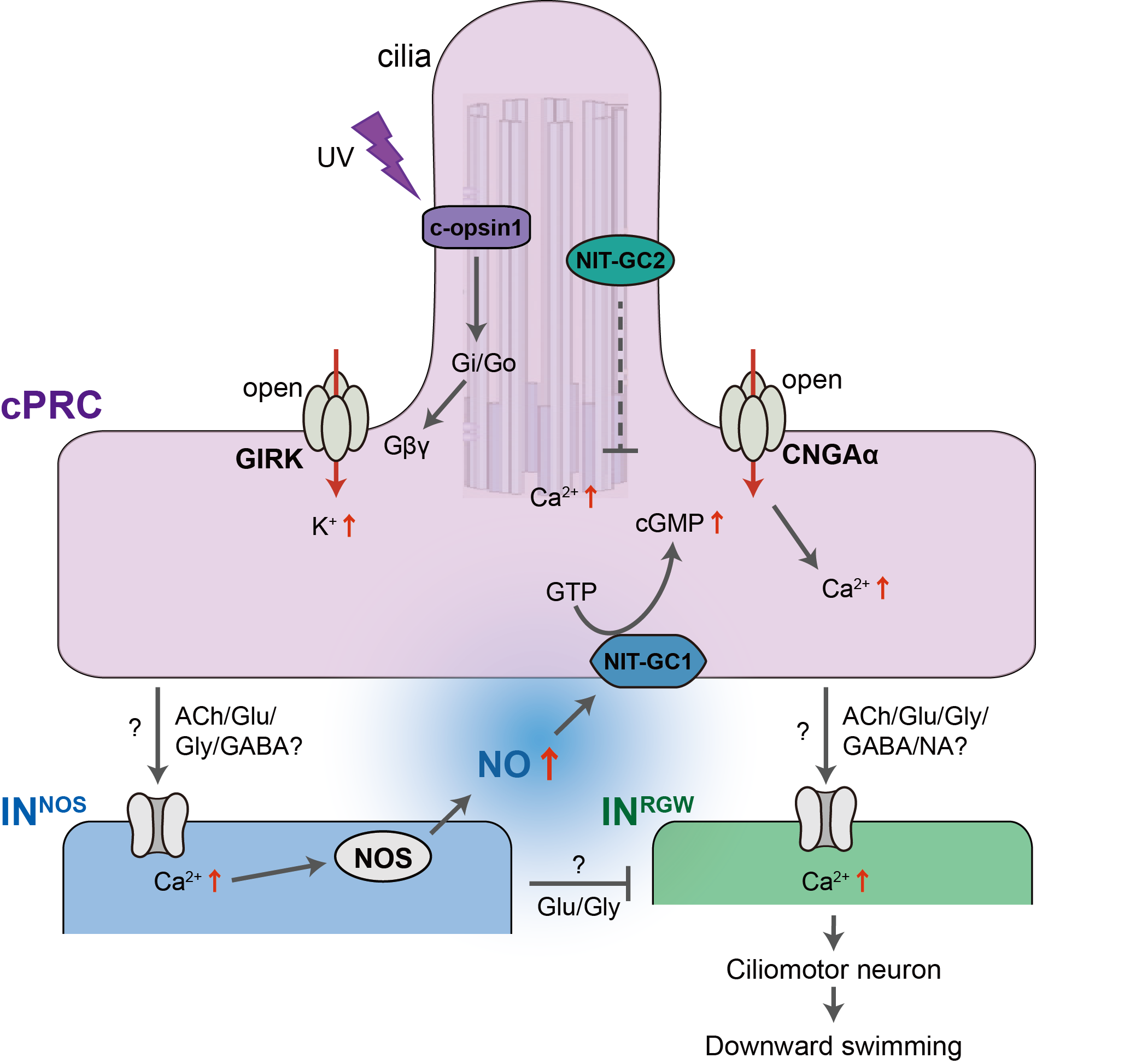
Circuitry of ciliary photoreceptors
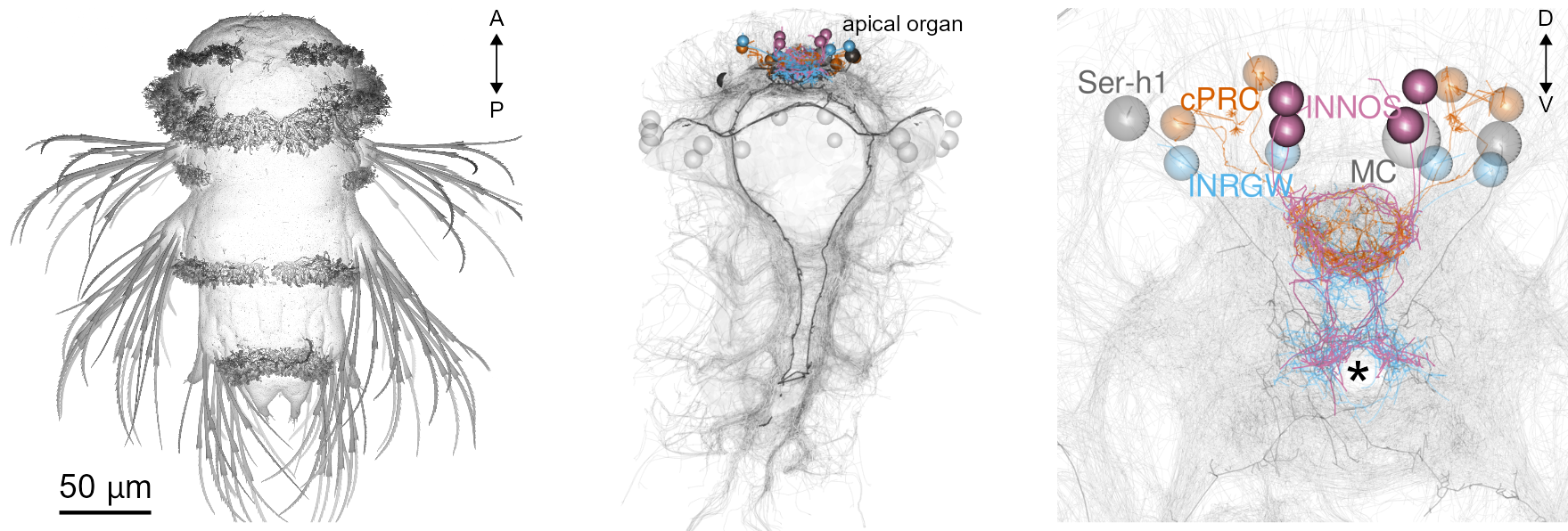
Circuitry of ciliary photoreceptors
Strong cPRC activation after UV exposure
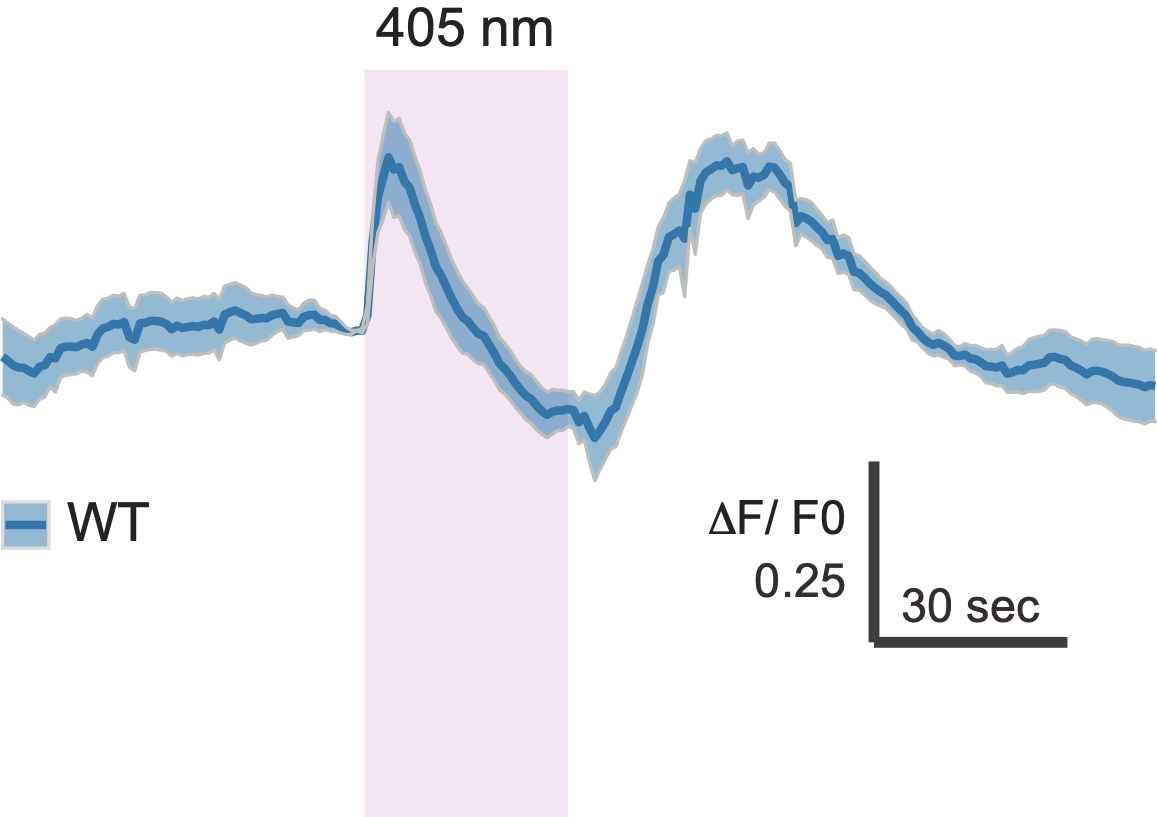
Single-cell data for cPRC cells and their interneurons
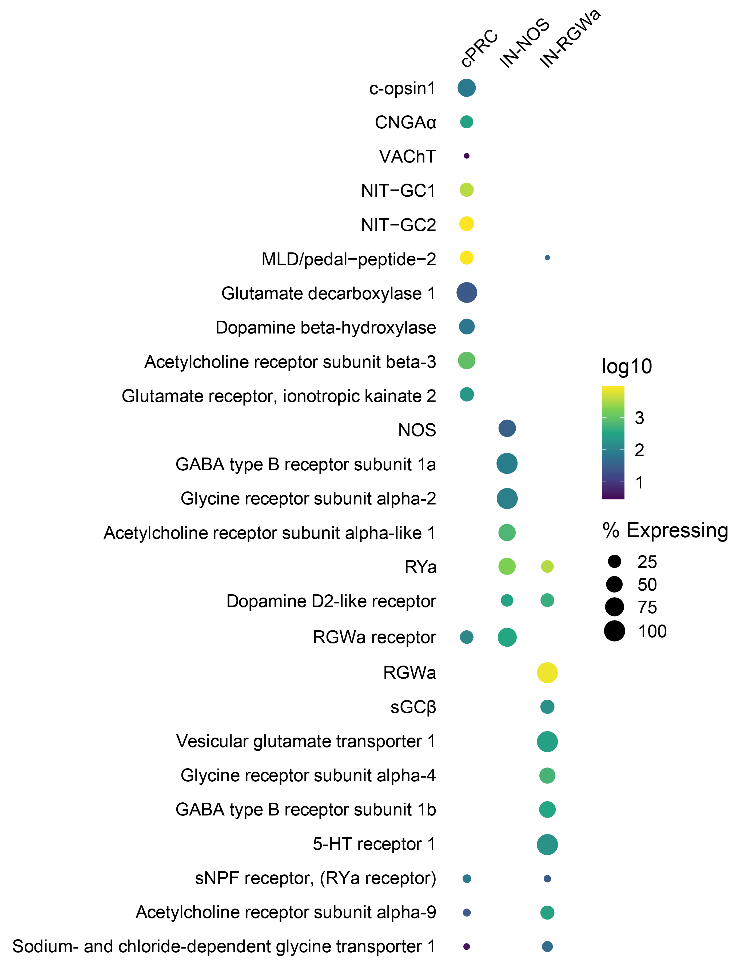 ::: aside Kei Jokura :::
::: aside Kei Jokura :::
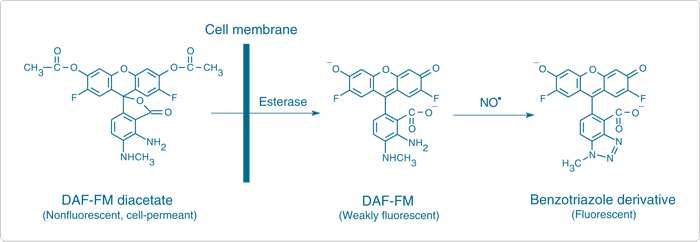
Nitric-oxyde synthase in postsynaptic interneurons
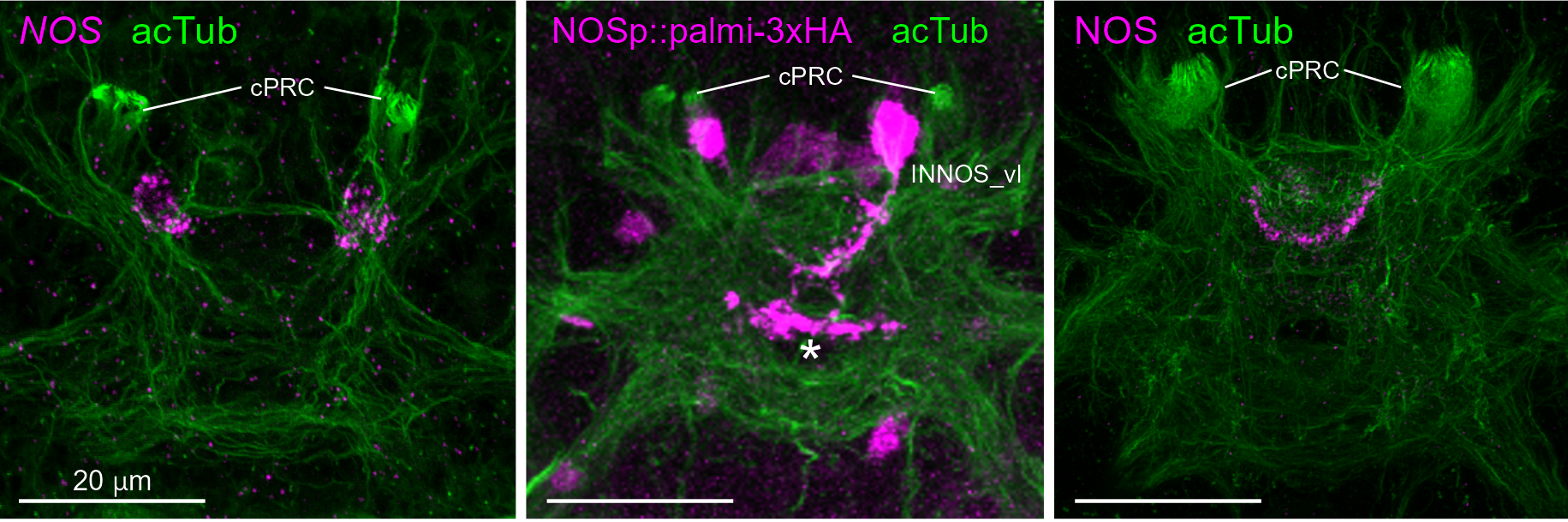
HCR
Transgenic labelling
NO is produced in the neuropil after UV stimulation
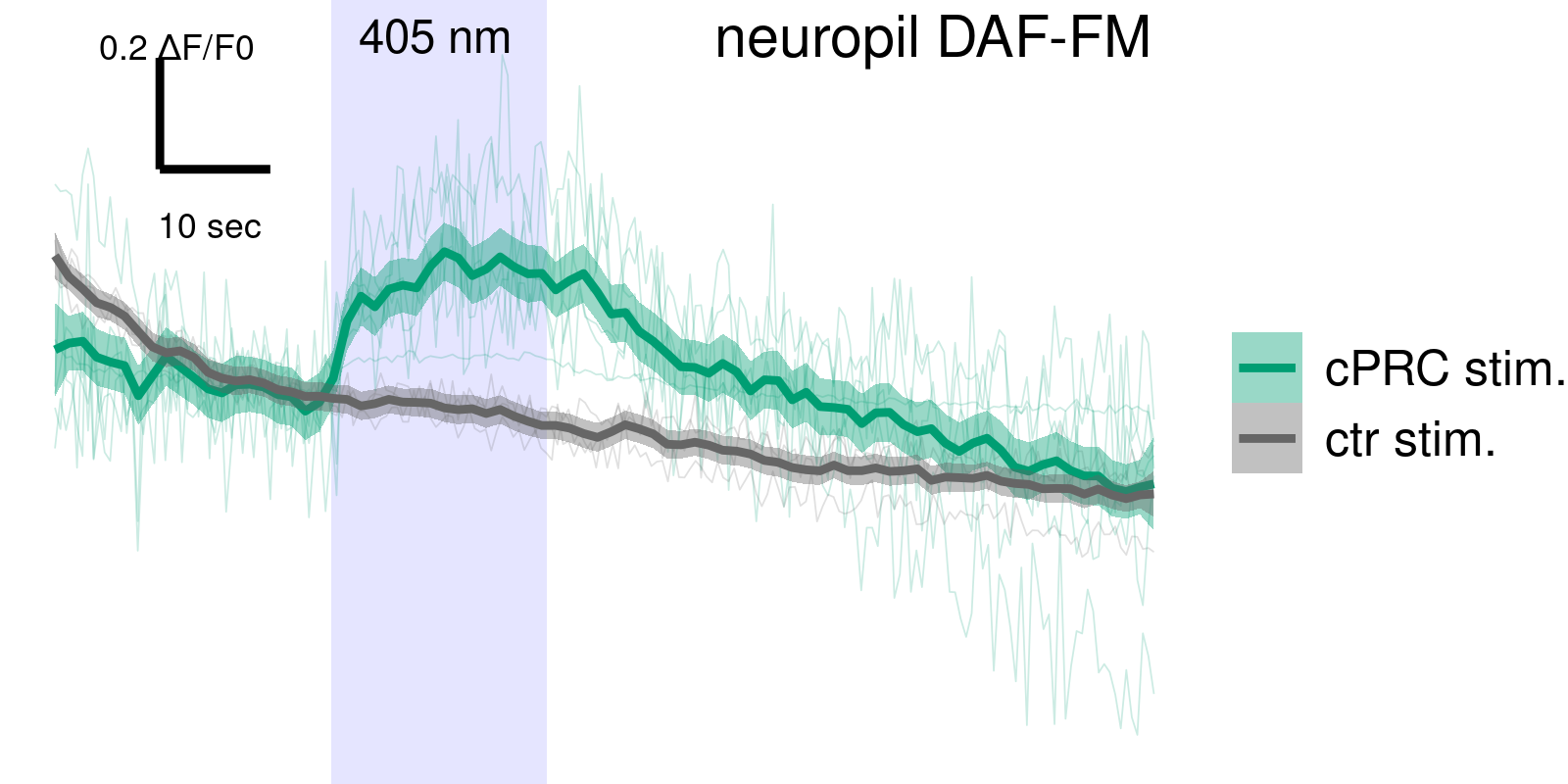
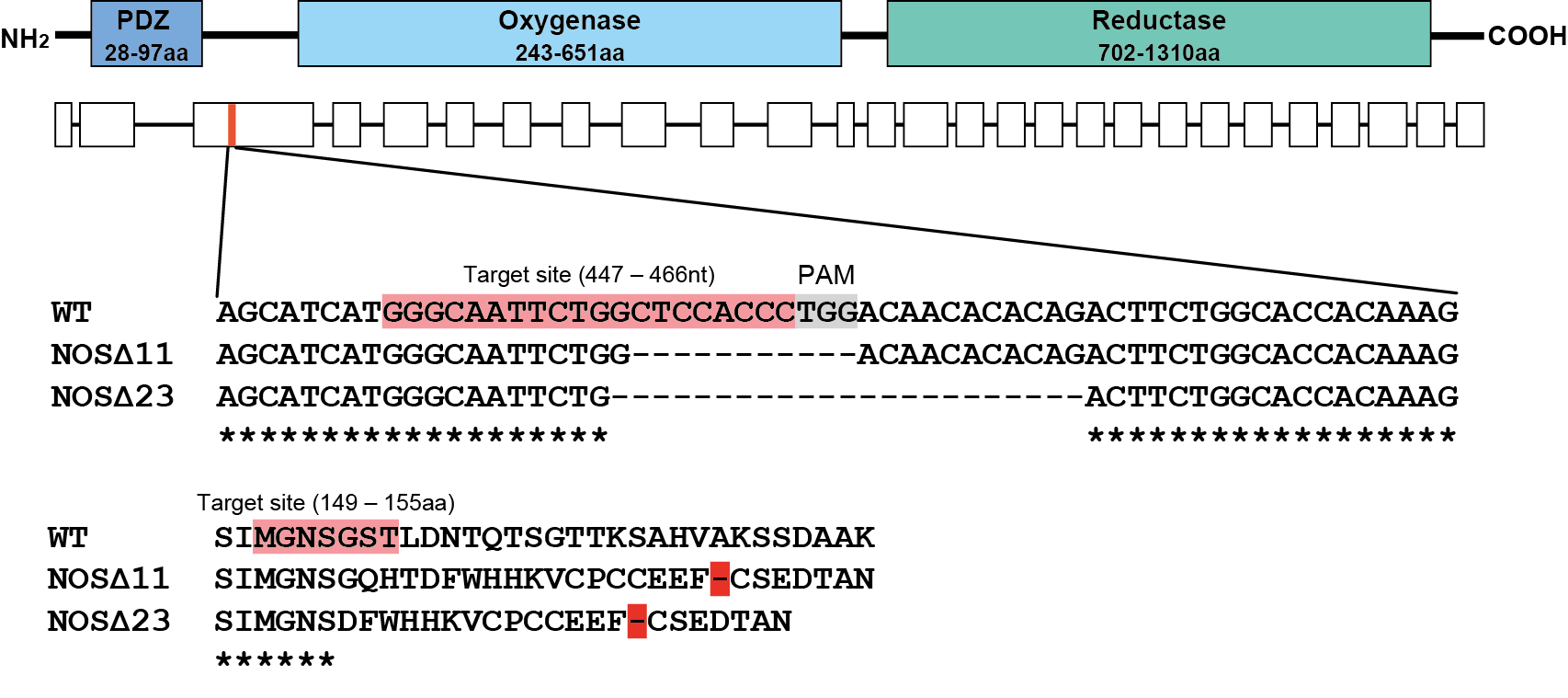
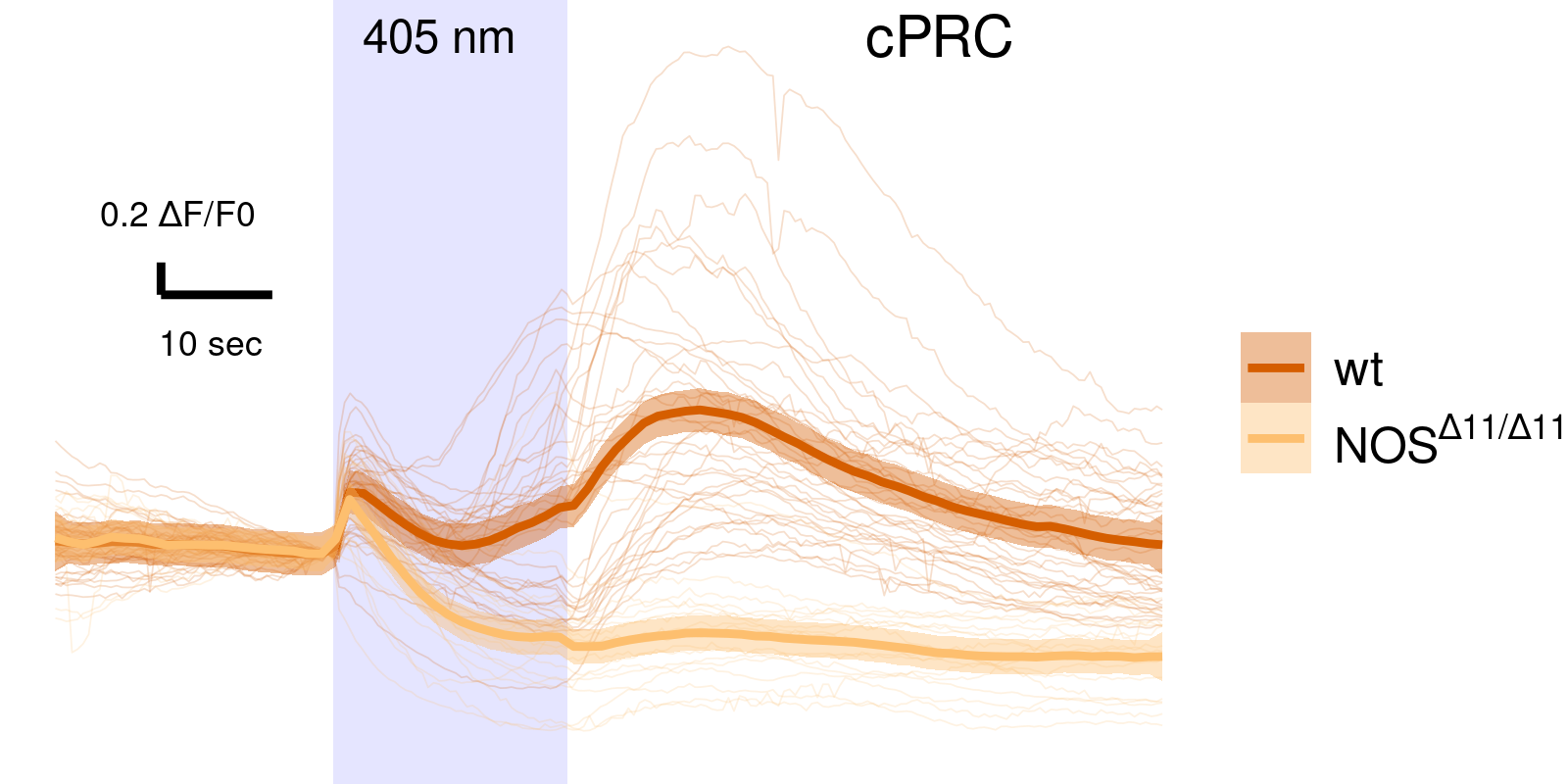
NOS mutants have altered cPRC response
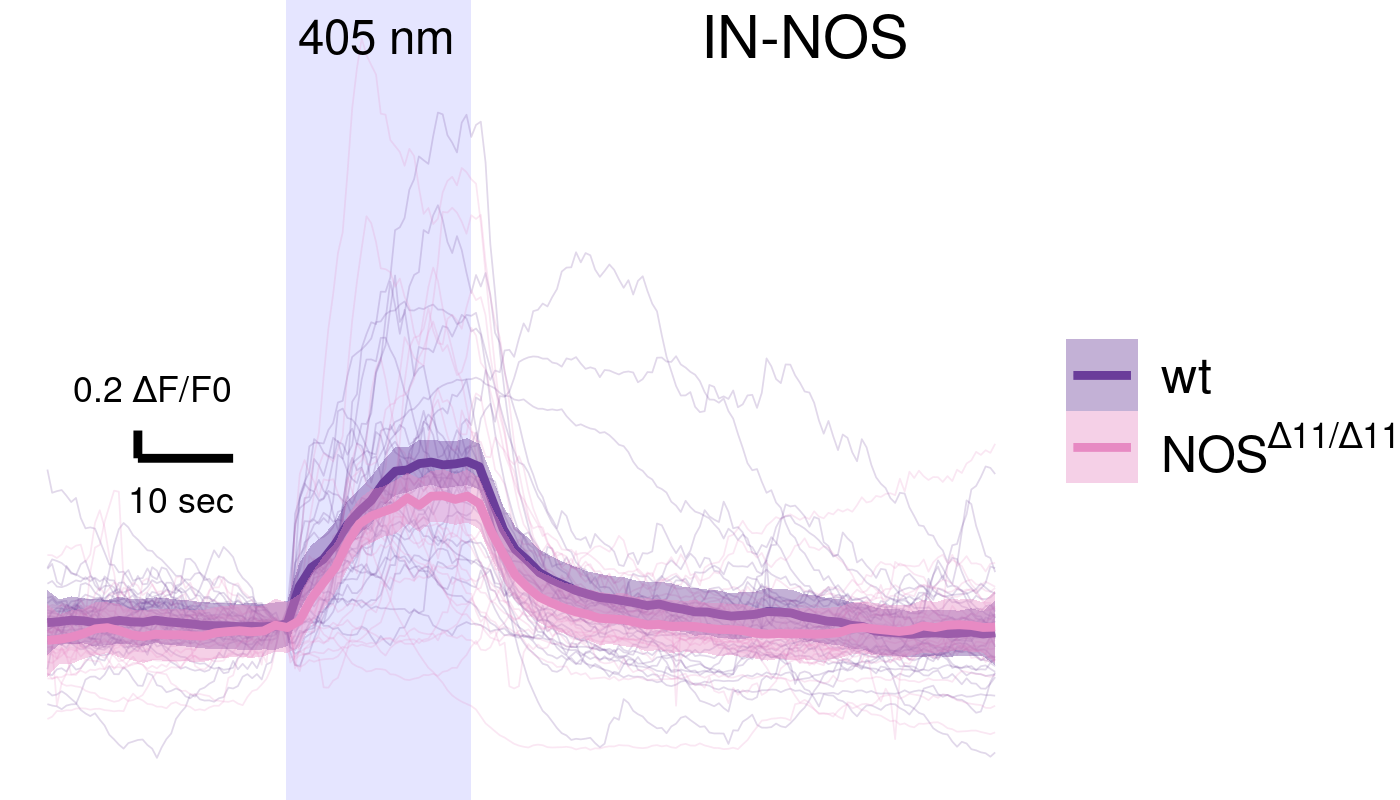
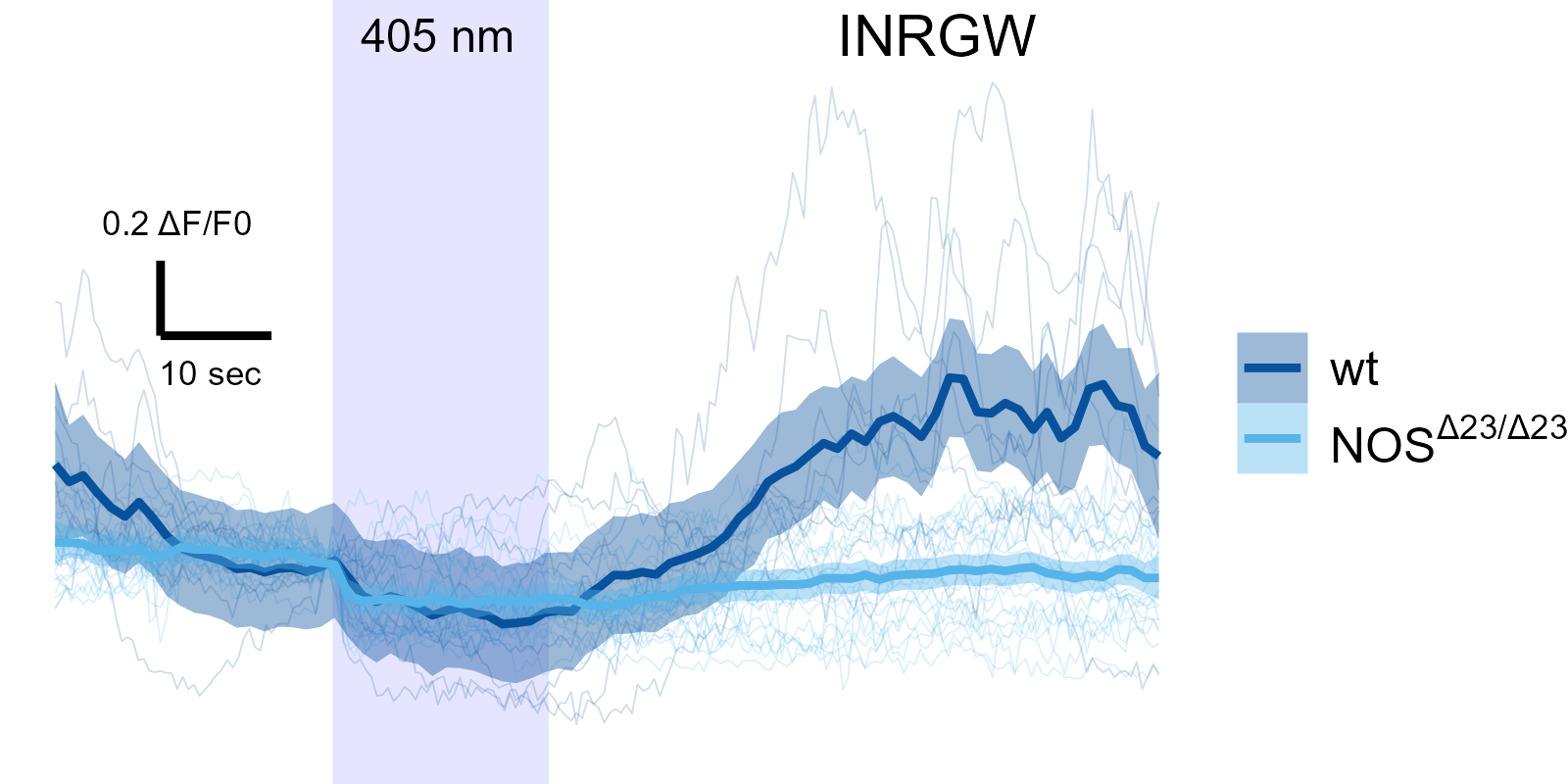
NOS mutants have altered INRGW and motoneuron response
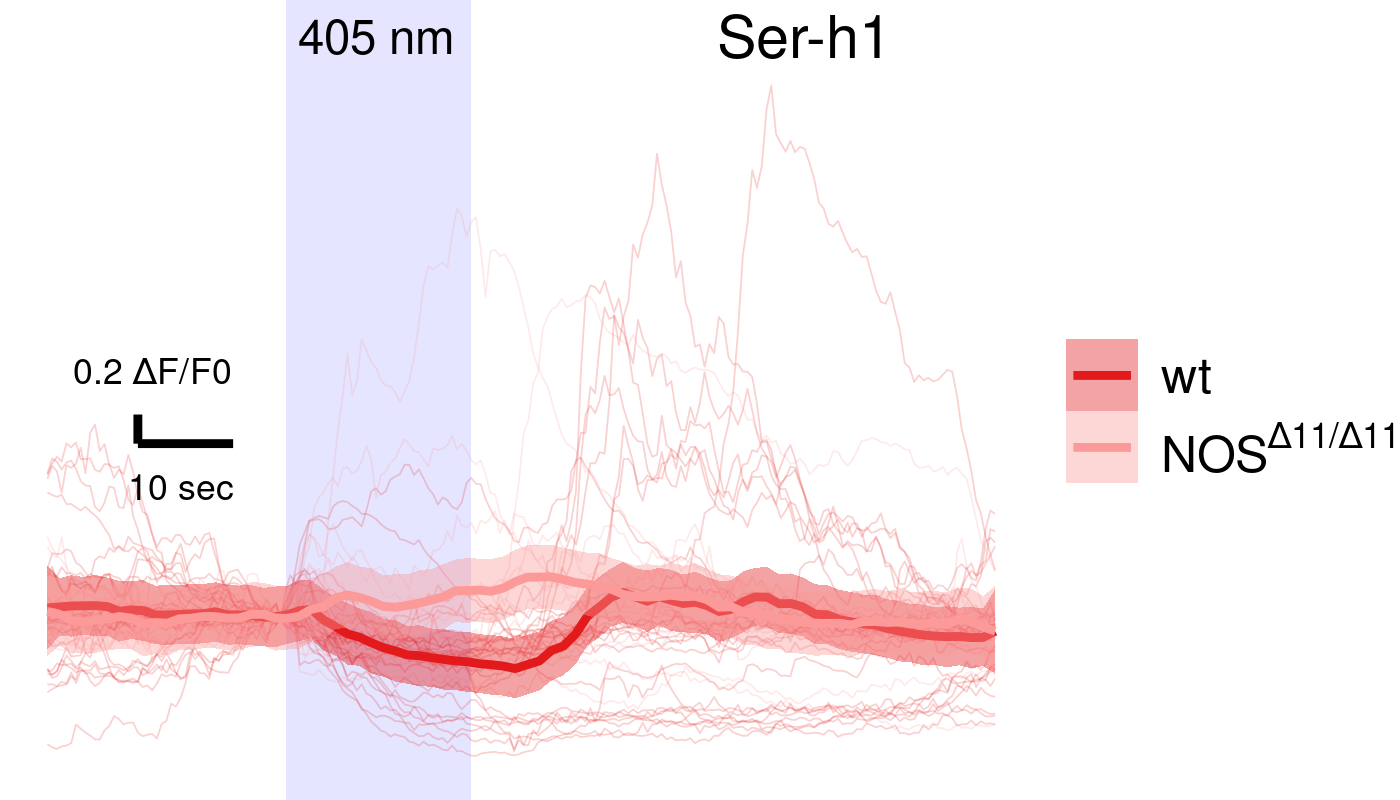

NOS mutants show defective UV avoidance
![]()
Two unusual guanylyl cyclases in the cPRCs
NIT-GC1 RNA
NIT-GC1 protein
NIT-GC2 RNA
NIT-GC2 protein
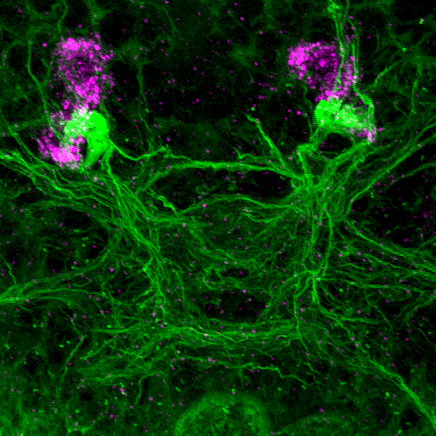
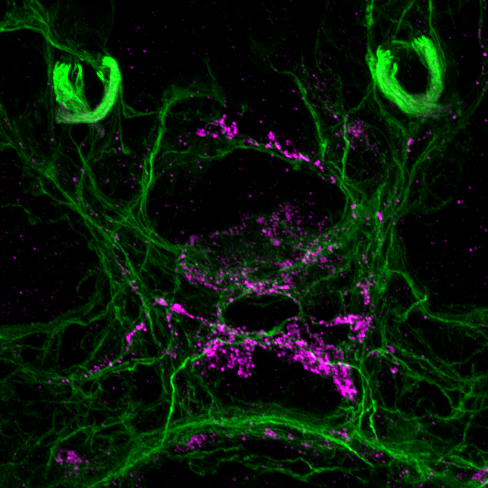
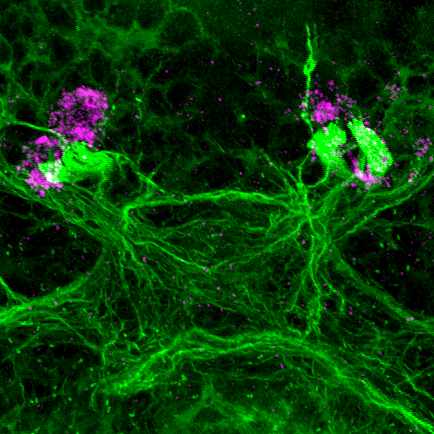
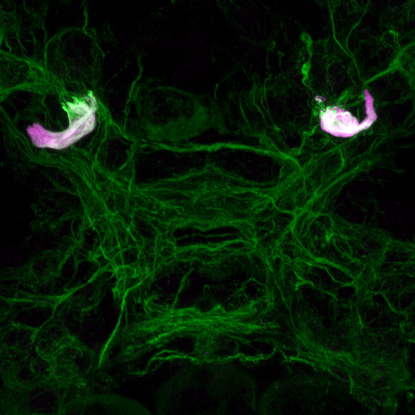
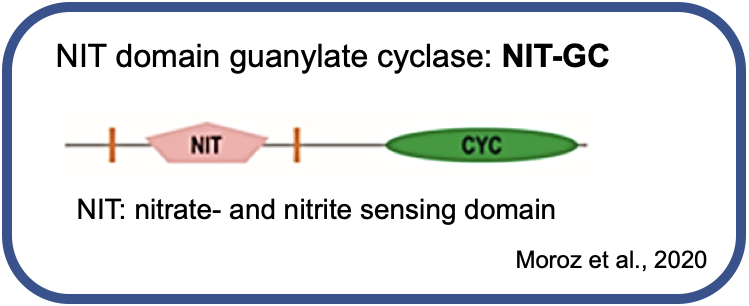
Two unusual guanylyl cyclases in the cPRCs

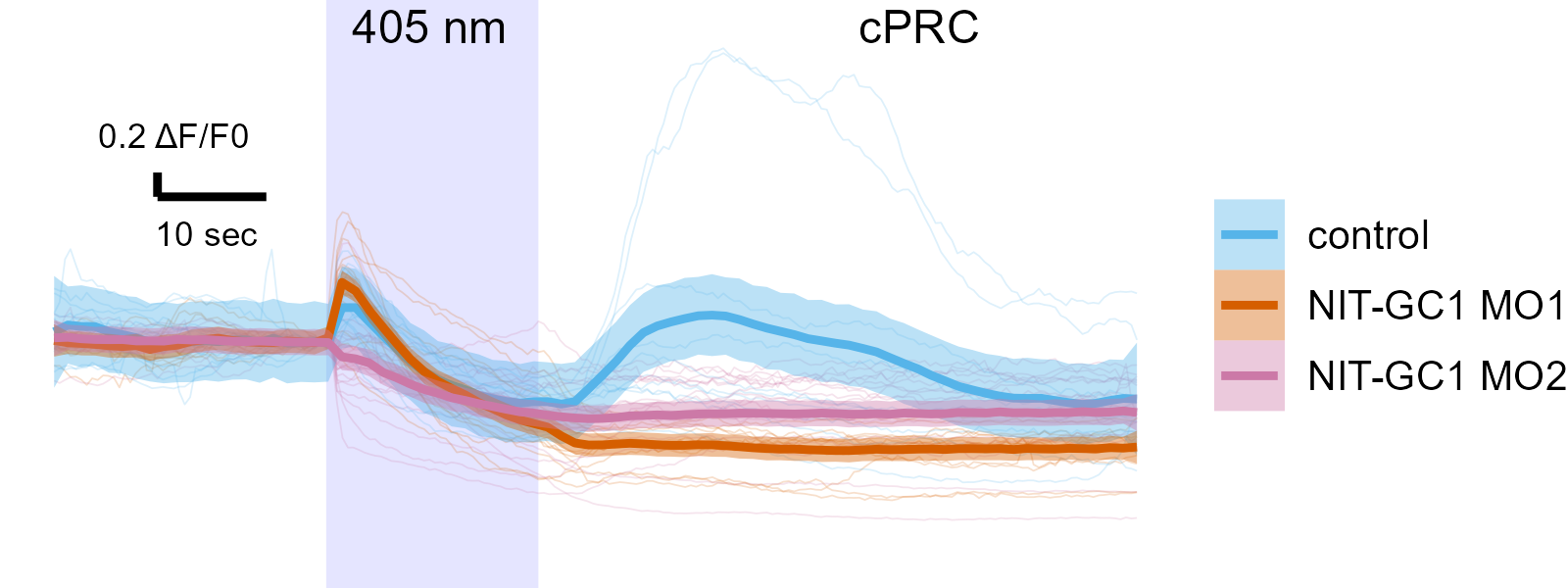
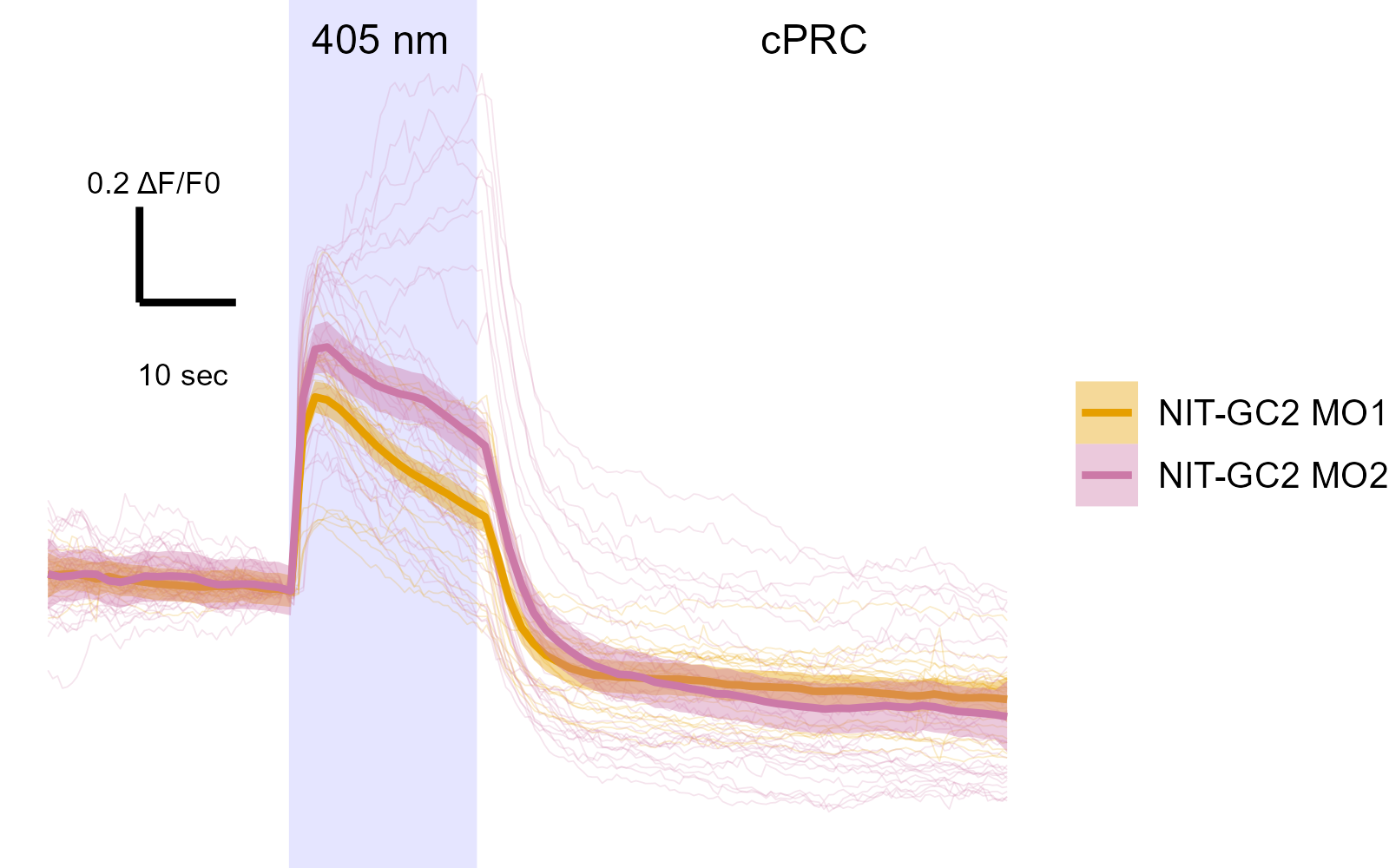
NIT-GC morphants have altered circuit activity

Mathematical modelling of the circuit
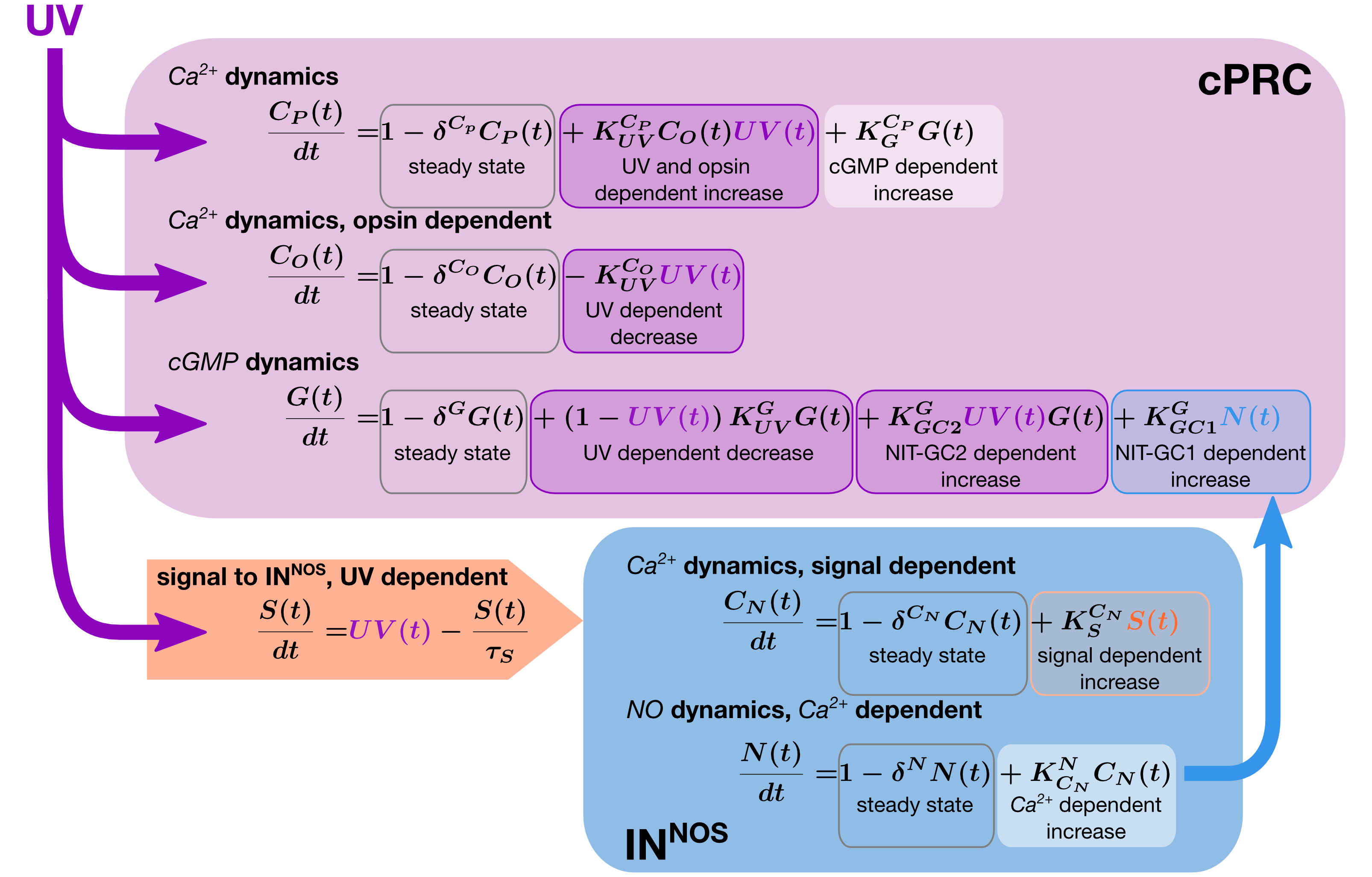
Model fitting
wild type
NOS-11
NOS-23
NIT-GC2 mo.
Integration and memory of UV exposure
Synaptic and chemical signalling work together

Acknowledgements
- Sanja Jasek
- Alexandra Kerbl
- Emily Savage
- Simone Wolters
- Lara Keweloh
- Kevin Urbansky
- Karel Mocaer
- David Hug
- Benedikt Dürr
- Ira Maegele
- Emelie Brodrick (Exeter)


Former lab members
- Kei Jokura, Luis A. Bezares-Calderón, Luis A. Yanez-Guerra, Victoria Moris, Daniel Thiel, Albina Asadulina, Cameron Hird, Adam Johnstone, Markus Conzelmann, Nadine Randel, Philipp Bauknecht, Martin Gühmann, Cristina Pineiro-Lopez, Nobuo Ueda, Aurora Panzera, Csaba Verasztó, Elizabeth Williams
